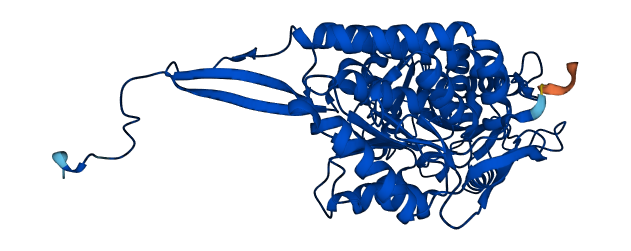
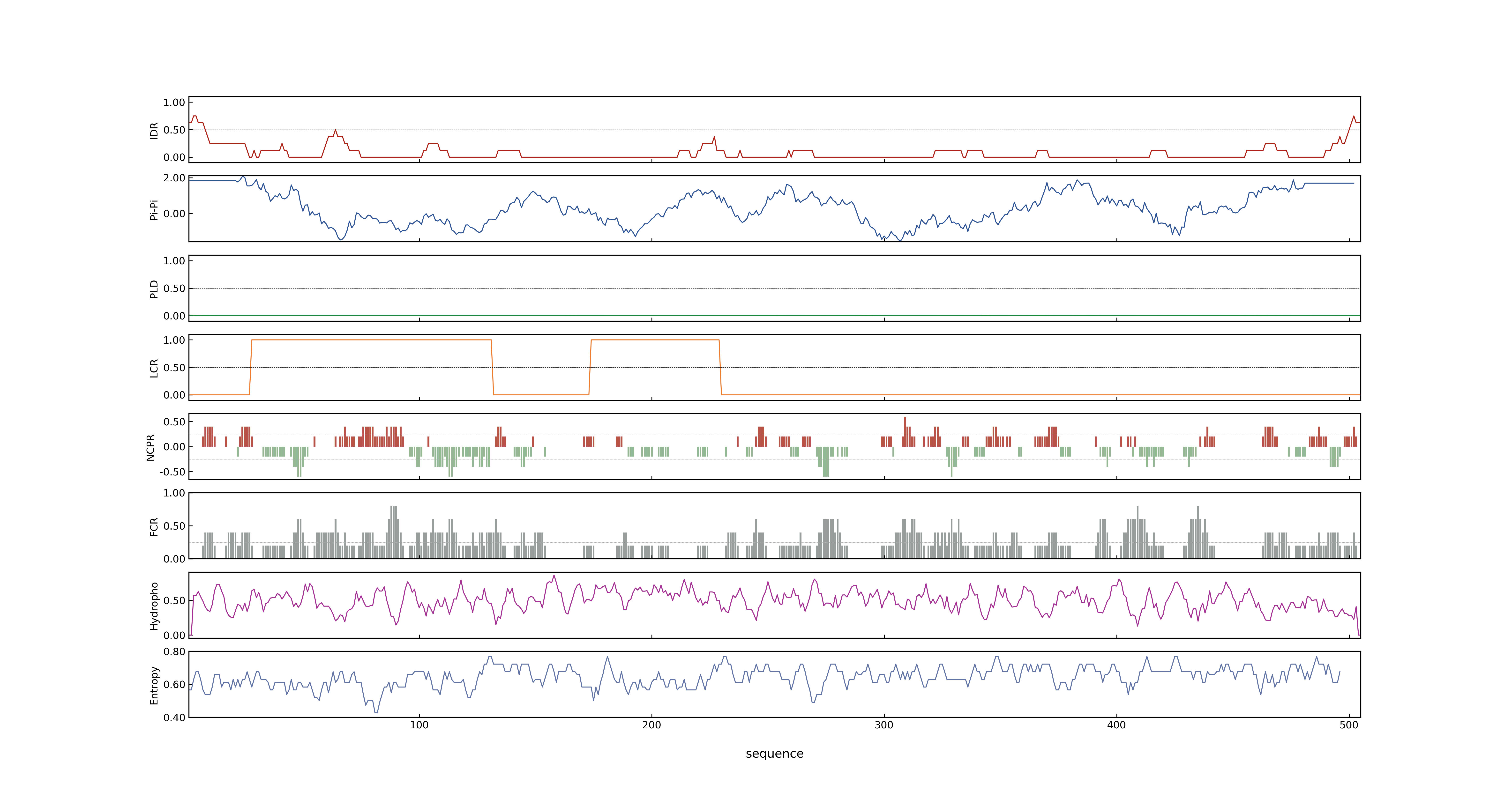
- Information
- Symbol: BAD1,OsBADH1
- MSU: LOC_Os04g39020
- RAPdb: Os04g0464200
- PSP score
- LOC_Os04g39020.1: 0.0033
- PLAAC score
- LOC_Os04g39020.1: 0
- pLDDT score
- 97.59
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os04g39020.1: 0.08557360
- MolPhase Result
- Publication
- OsBADH1 is possibly involved in acetaldehyde oxidation in rice plant peroxisomes, 2009, FEBS Lett.
- Dissecting substrate specificity of two rice BADH isoforms: Enzyme kinetics, docking and molecular dynamics simulation studies, 2012, Biochimie.
- Genetic manipulation of Japonica rice using the OsBADH1 gene from Indica rice to improve salinity tolerance, 2010, Plant Cell, Tissue and Organ Culture (PCTOC).
- Expression of OsBADH1 gene in Indica rice Oryza sativaL. in correlation with salt, plasmolysis, temperature and light stresses , 2011, Plant Omics.
- Inactivation of an aminoaldehyde dehydrogenase is responsible for fragrance in rice, 2008, Plant Mol Biol.
- Expression of Indica rice OsBADH1 gene under salinity stress in transgenic tobacco, 2009, Plant Biotechnology Reports.
- RNAi-directed downregulation of betaine aldehyde dehydrogenase 1 OsBADH1 results in decreased stress tolerance and increased oxidative markers without affecting glycine betaine biosynthesis in rice Oryza sativa., 2014, Plant Mol Biol.
- Resequencing Reveals Different Domestication Rate for BADH1 and BADH2 in Rice Oryza sativa., 2015, PLoS One.
- Genbank accession number
- Key message
- Northern blot analysis revealed that salt-tolerant in each rice cultivar is correlated to the expression level of OsBADH1 mRNA
- Moreover, these results suggest that the expression of OsBADH1 gene in response to salt stress could be magnified under high light conditions
- Interestingly, the effect of salt stress on the expression of OsBADH1 gene was alleviated by CO2 enrichment
- Expression of OsBADH1 gene in Indica rice (Oryza sativaL.) in correlation with salt, plasmolysis, temperature and light stresses
- We previously found that the expression of betaine aldehyde dehydrogenase 1 gene (OsBADH1), encoding a key enzyme for glycine betaine biosynthesis pathway, showed close correlation with salt tolerance of rice
- In this study, the expression of the OsBADH1 gene in transgenic tobacco was investigated in response to salt stress using a transgenic approach
- The expression studies showed that OsBADH1 can be induced by a variety of environmental factors such as salinity, drought, cold, heat, light intensity and CO2 concentration
- The results demonstrated that the OsBADH1 mRNA expression was up-regulated by salinity, drought, cold and high light intensity but down-regulated by CO2 enrichment and heat stress
- The primary response of OsBADH1 gene expression was induced within 24 h after salinity, cold or drought stress treatment
- Expression of Indica rice OsBADH1 gene under salinity stress in transgenic tobacco
- Japonica rice (salt-sensitive) was genetically engineered to enhance salt tolerance by introducing the OsBADH1 gene from Indica rice (salt-tolerant), which is a GB accumulator
- Genetic manipulation of Japonica rice using the OsBADH1 gene from Indica rice to improve salinity tolerance
- The accumulation of OsBADH1 mRNA decreases following submergence treatment, but quickly recovers after re-aeration
- Transgenic rice lines downregulating OsBADH1 exhibited remarkably reduced tolerance to NaCl, drought and cold stresses
- In this study, we demonstrated a pivotal role of OsBADH1 in stress tolerance without altering GB biosynthesis capacity, using the RNA interference (RNAi) technique
- Connection
- BAD1~OsBADH1, OsBADH2~fgr, OsBADH1 is possibly involved in acetaldehyde oxidation in rice plant peroxisomes, We found that OsBADH1 catalyzes the oxidation of acetaldehyde efficiently, while the activity of OsBADH2 is extremely low
- BAD1~OsBADH1, OsBADH2~fgr, Dissecting substrate specificity of two rice BADH isoforms: Enzyme kinetics, docking and molecular dynamics simulation studies, Fragrance rice (Oryza sativa) contains two isoforms of BADH, named OsBADH1 and OsBADH2
- BAD1~OsBADH1, OsBADH2~fgr, Dissecting substrate specificity of two rice BADH isoforms: Enzyme kinetics, docking and molecular dynamics simulation studies, OsBADH1 is implicated in acetaldehyde oxidation in rice plant peroxisomes, while the non-functional OsBADH2 is believed to be involved in the accumulation of 2-acetyl-1-pyrroline, the major compound of aroma in fragrance rice
- BAD1~OsBADH1, OsBADH2~fgr, Dissecting substrate specificity of two rice BADH isoforms: Enzyme kinetics, docking and molecular dynamics simulation studies, Consistent with our previous study, kinetics data indicated that the enzymes catalyze the oxidation of GAB-ald more efficiently than Bet-ald and the OsBADH1 W172F and OsBADH2 W170F mutants displayed a higher catalytic efficiency towards GAB-ald
- BAD1~OsBADH1, OsBADH2~fgr, Dissecting substrate specificity of two rice BADH isoforms: Enzyme kinetics, docking and molecular dynamics simulation studies, The amino acid residues, E262, L263, C296 and W461 of OsBADH1 and E260, L261, C294 and W459 of OsBADH2 located within 5 A of the OsBADH active site mainly interacted with GAB-ald forming strong hydrogen bonds in both OsBADH isoforms
- BAD1~OsBADH1, OsBADH2~fgr, Dissecting substrate specificity of two rice BADH isoforms: Enzyme kinetics, docking and molecular dynamics simulation studies, Residues W163, N164, Q294, C296 and F397 of OsBADH1-Bet-ald and Y163, M167, W170, E260, S295 and C453 of OsBADH2-Bet-ald formed the main interaction sites while E260 showed an interaction energy of -14
- BAD1~OsBADH1, OsBADH2~fgr, Dissecting substrate specificity of two rice BADH isoforms: Enzyme kinetics, docking and molecular dynamics simulation studies, Unconserved A290 in OsBADH1 and W288 in OsBADH2 appeared to be important for substrate recognition similar to that observed in PsAMADHs
Prev Next