- Information
- Symbol: DEP1,DN1,qPE9-1,OsDEP1
- MSU: LOC_Os09g26999
- RAPdb: Os09g0441900
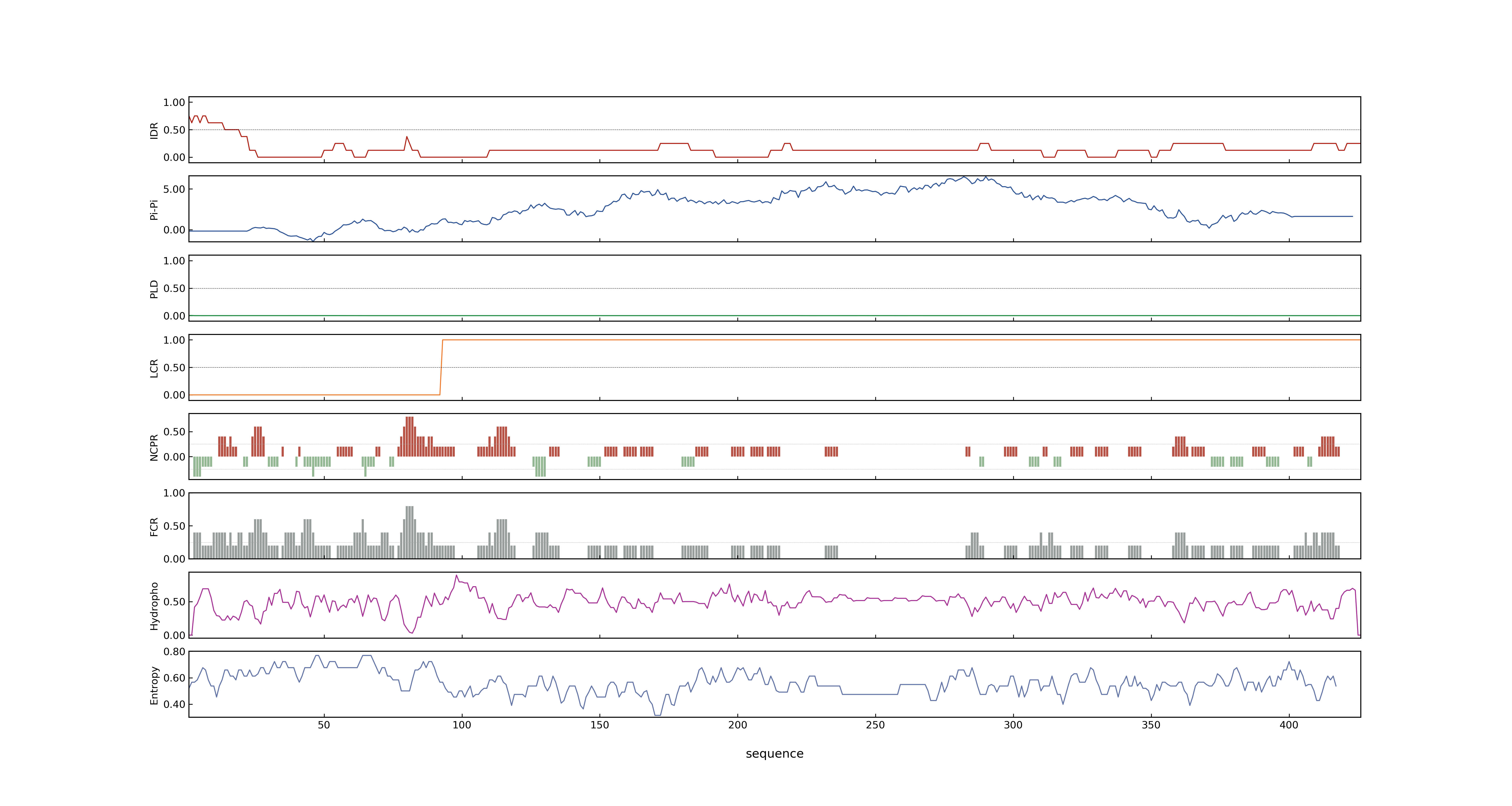
- PSP score
- LOC_Os09g26999.2: 0.0036
- LOC_Os09g26999.1: 0.961
- LOC_Os09g26999.3: 0.0065
- PLAAC score
- LOC_Os09g26999.2: 0
- LOC_Os09g26999.1: 0
- LOC_Os09g26999.3: 0
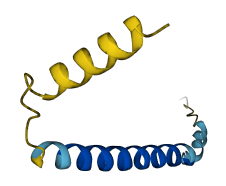
- pLDDT score
- 76.93
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os09g26999.1: 0.92711019
- LOC_Os09g26999.2: 0.20422793
- LOC_Os09g26999.3: 0.00094156
- MolPhase Result
- Publication
- Introgression of qPE9-1 allele, conferring the panicle erectness, leads to the decrease of grain yield per plant in japonica rice Oryza sativa L., 2011, J Genet Genomics.
- Identification and characterization of a major QTL responsible for erect panicle trait in japonica rice Oryza sativa L., 2007, Theor Appl Genet.
- Deletion in a quantitative trait gene qPE9-1 associated with panicle erectness improves plant architecture during rice domestication, 2009, Genetics.
- A loss-of-function mutation of rice DENSE PANICLE 1 causes semi-dwarfness and slightly increased number of spikelets, 2011, Breeding Science.
- Rice DEP1, encoding a highly cysteine-rich G protein gamma subunit, confers cadmium tolerance on yeast cells and plants, 2013, J Exp Bot.
- Natural variation at the DEP1 locus enhances grain yield in rice, 2009, Nat Genet.
- Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture, 2013, Plant Cell.
- The plant-specific G protein gamma subunit AGG3 influences organ size and shape in Arabidopsis thaliana, 2012, New Phytol.
- Variations in DENSE AND ERECT PANICLE 1 DEP1 contribute to the diversity of the panicle trait in high-yielding japonica rice varieties in northern China., 2016, Breed Sci.
- The DENSE AND ERECT PANICLE 1 DEP1 gene offering the potential in the breeding of high-yielding rice., 2016, Breed Sci.
-
Genbank accession number
- Key message
- The expression level of OsCKX2 in the shoot apex of Dn1-1 plants is similar to that in the wild type, indicating that OsCKX2 does not contribute to an increased number of spikelets
- DN1 is allelic to DENSE AND ERECT PANICLE 1 (DEP1) (=qPE(9-1))
- Recently, qPE9-1 has been successfully cloned; however, the genetic effect on grain yield per plant of the erect panicle allele qPE9-1 is controversial yet
- The comparison of agronomic traits between the NILs showed that, when qpe9-1 was replaced by qPE9-1, the panicle architecture was changed from drooping to erect; moreover, the panicle length, plant height, 1000-grain weight and the tillers were significantly decreased, consequently resulting in the dramatic decrease of grain yield per plant by 30%
- Therefore, we concluded that the qPE9-1 was a major factor controlling panicle architecture, and qPE9-1 had pleiotropic nature, with negative effects on grain yield per plant
- Introgression of qPE9-1 allele, conferring the panicle erectness, leads to the decrease of grain yield per plant in japonica rice (Oryza sativa L.)
- The qPE9-1 gene has been proved to be widely used in high-yield rice cultivar developments, conferring erect panicle character in japonica rice
- In the present study, a drooping panicle parent Nongken 57, carrying qpe9-1 allele, was used as recurrent parent to successively backcross to a typical erect panicle line from the double haploid (DH) population (Wuyunjing 8/Nongken 57), which was previously shown to carry qPE9-1 allele
- This result strongly suggests that the erect panicle allele qPE9-1 should be used together with other favorable genes in the high-yield breeding practice
- A comparison of the Dn1-1 and Dn1-3 alleles suggests that the N-terminal region of DN1 contains a coiled-coil domain and a nuclear localization signal that might be responsible for semi-dwarfness
- A rice cDNA, OsDEP1, encoding a highly cysteine (Cys)-rich G protein gamma subunit, was initially identified as it conferred cadmium (Cd) tolerance on yeast cells
- Rice DEP1, encoding a highly cysteine-rich G protein gamma subunit, confers cadmium tolerance on yeast cells and plants
- Natural variation at the DEP1 locus enhances grain yield in rice
- In addition, the qPE9-1 locus regulates panicle and grain length, grain weight, and consequently grain yield
- Dn1-1 plants have normal sensitivity to gibberellin, brassinolide, and kinetin, and we observed no genetic epistasis with brassinolide-related mutants, suggesting that DN1 does not function in the signaling pathways of these phytohormones
- Here we report the map-based cloning of a major quantitative trait locus, qPE9-1, which plays an integral role in regulation of rice plant architecture including panicle erectness
- Phenotypic comparisons of a set of near-isogenic lines and transgenic lines reveal that the functional allele (qPE9-1) results in drooping panicles, and the loss-of-function mutation (qpe9-1) leads to more erect panicles
- We propose that the panicle erectness trait resulted from a natural random loss-of-function mutation for the qPE9-1 gene and has subsequently been the target of artificial selection during japonica rice breeding
- Deletion in a quantitative trait gene qPE9-1 associated with panicle erectness improves plant architecture during rice domestication
- In addition, we found that H90, the nearest marker to qPE9-1, used for genotyping 38 cultivars with extremely erect and drooping panicles, segregated in agreement with PC, suggesting the H90 product was possibly part of the qPE9-1 gene or closely related to it
- We show that the rice DENSE PANICLE 1 (DN1) mutant allele Dn1-1 causes both of these characteristics and that Dn1-1 is a loss-of-function mutation
- These data demonstrated that H90 could be used for marker-aided selection for the PE trait in breeding and in the cloning of qPE9-1
- In order to improve the genetic diversity of DEP1, we used a rice germplasm collection of 72 high yielding japonica rice varieties to analyze the contribution of DEP1 to the panicle traits
- The SNP (G/C) at the promoter region will contribute to the flexible application of DEP1 in rice breeding
- DEP1 is a G protein gamma subunit that is involved in the regulation of erect panicle, number of grains per panicle, nitrogen uptake, and stress-tolerance through the G protein signal pathway
- Here we review the development of erect panicle rice varieties, DEP1 alleles and regulatory network, and its physiological and morphological functions
- Additionally, the further increasing the yield potential of erect-panicle super-rice, and the development of molecular designing breeding for indica-japonica hybrid rice with the dep1 gene are also prospected
- Connection
- DEP1~DN1~qPE9-1~OsDEP1, Gn1a~OsCKX2, A loss-of-function mutation of rice DENSE PANICLE 1 causes semi-dwarfness and slightly increased number of spikelets, The expression level of OsCKX2 in the shoot apex of Dn1-1 plants is similar to that in the wild type, indicating that OsCKX2 does not contribute to an increased number of spikelets
- DEP1~DN1~qPE9-1~OsDEP1, RGB1, Rice G-protein subunits qPE9-1 and RGB1 play distinct roles in abscisic acid responses and drought adaptation., Rice G-protein subunits qPE9-1 and RGB1 play distinct roles in abscisic acid responses and drought adaptation.
- DEP1~DN1~qPE9-1~OsDEP1, RGB1, Rice G-protein subunits qPE9-1 and RGB1 play distinct roles in abscisic acid responses and drought adaptation., Furthermore, the results suggested that qPE9-1 negatively regulates the ABA response by suppressing the expression of key transcription factors involved in ABA and stress responses, while RGB1 positively regulates ABA biosynthesis by upregulating NCED gene expression under both normal and drought stress conditions
- DEP1~DN1~qPE9-1~OsDEP1, RGB1, Rice G-protein subunits qPE9-1 and RGB1 play distinct roles in abscisic acid responses and drought adaptation., Taken together, it is proposed that RGB1 is a positive regulator of the ABA response and drought adaption in rice plants, whereas qPE9-1 is modulated by RGB1 and functions as a negative regulator in the ABA-dependent drought-stress responses
- DEP1~DN1~qPE9-1~OsDEP1, RGG1, Function of heterotrimeric G-protein gamma subunit RGG1 in providing salinity stress tolerance in rice by elevating detoxification of ROS., In salinity-stressed RGG1 transgenic lines, the transcript levels of RGG2, RGB, RGA, DEP1, and GS3 also increased in addition to RGG1
Prev Next