- Information
- Symbol: DTH2,OsCOL9,OsCCT08
- MSU: LOC_Os02g49230
- RAPdb: Os02g0724000
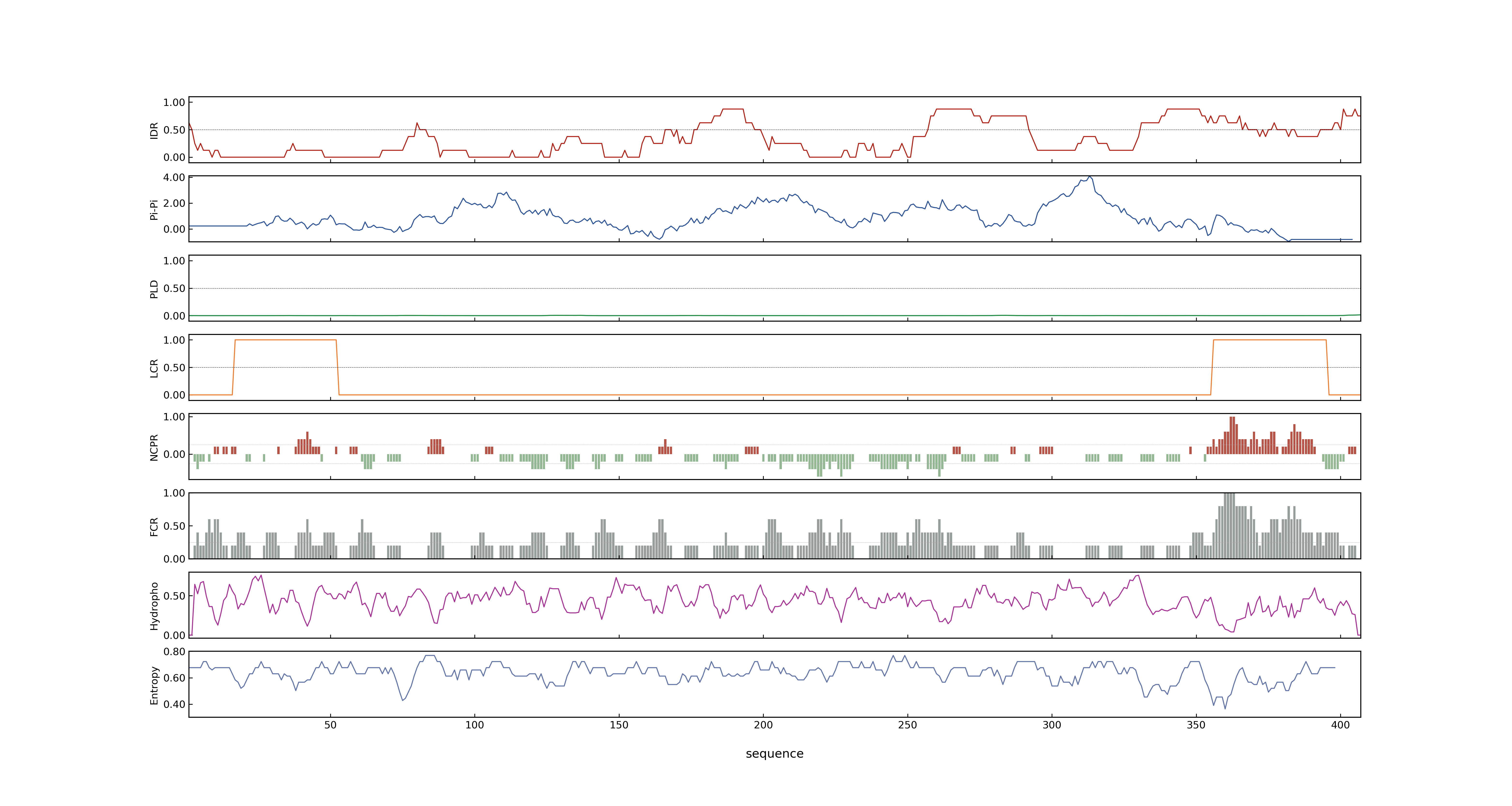
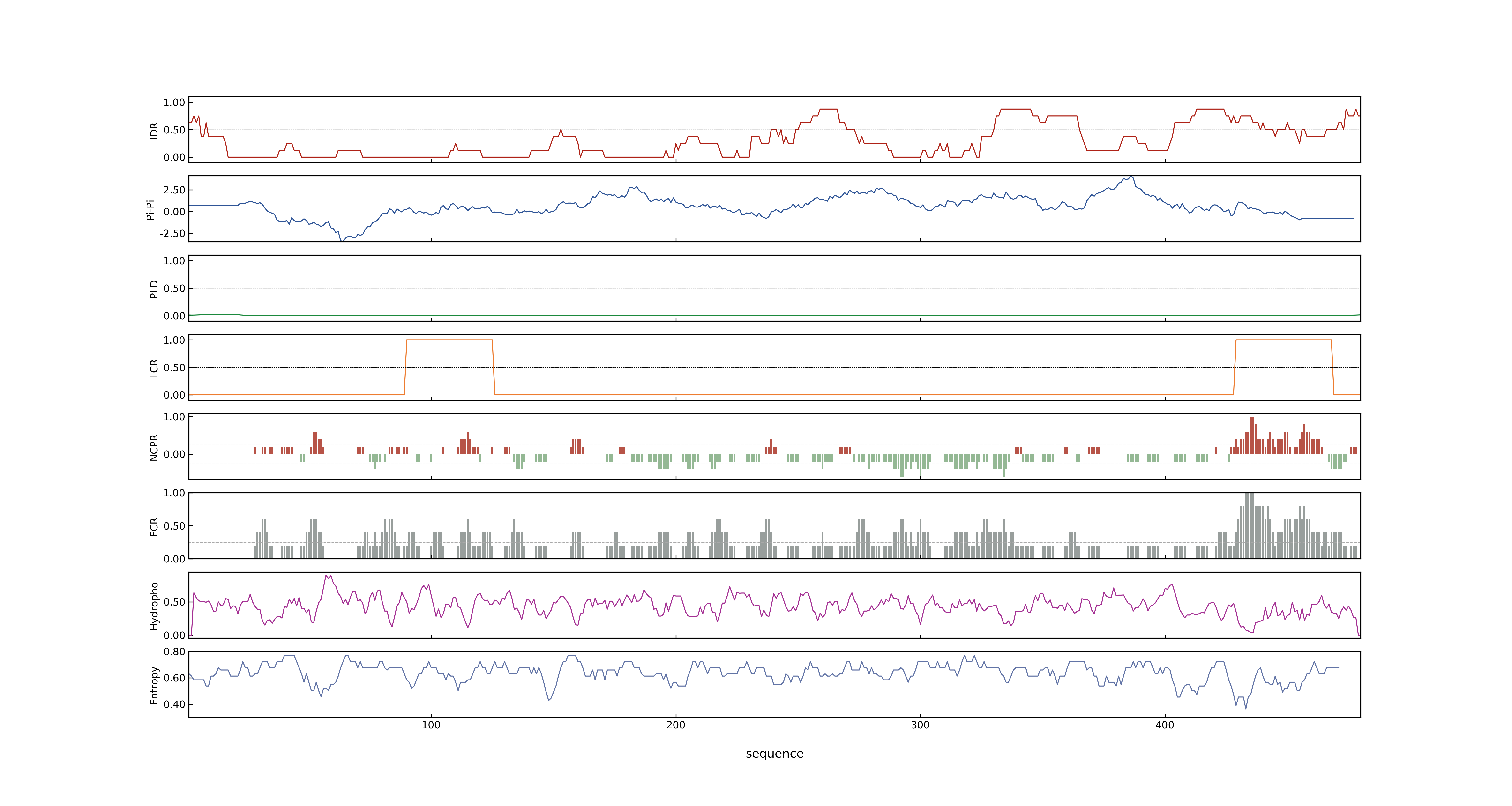
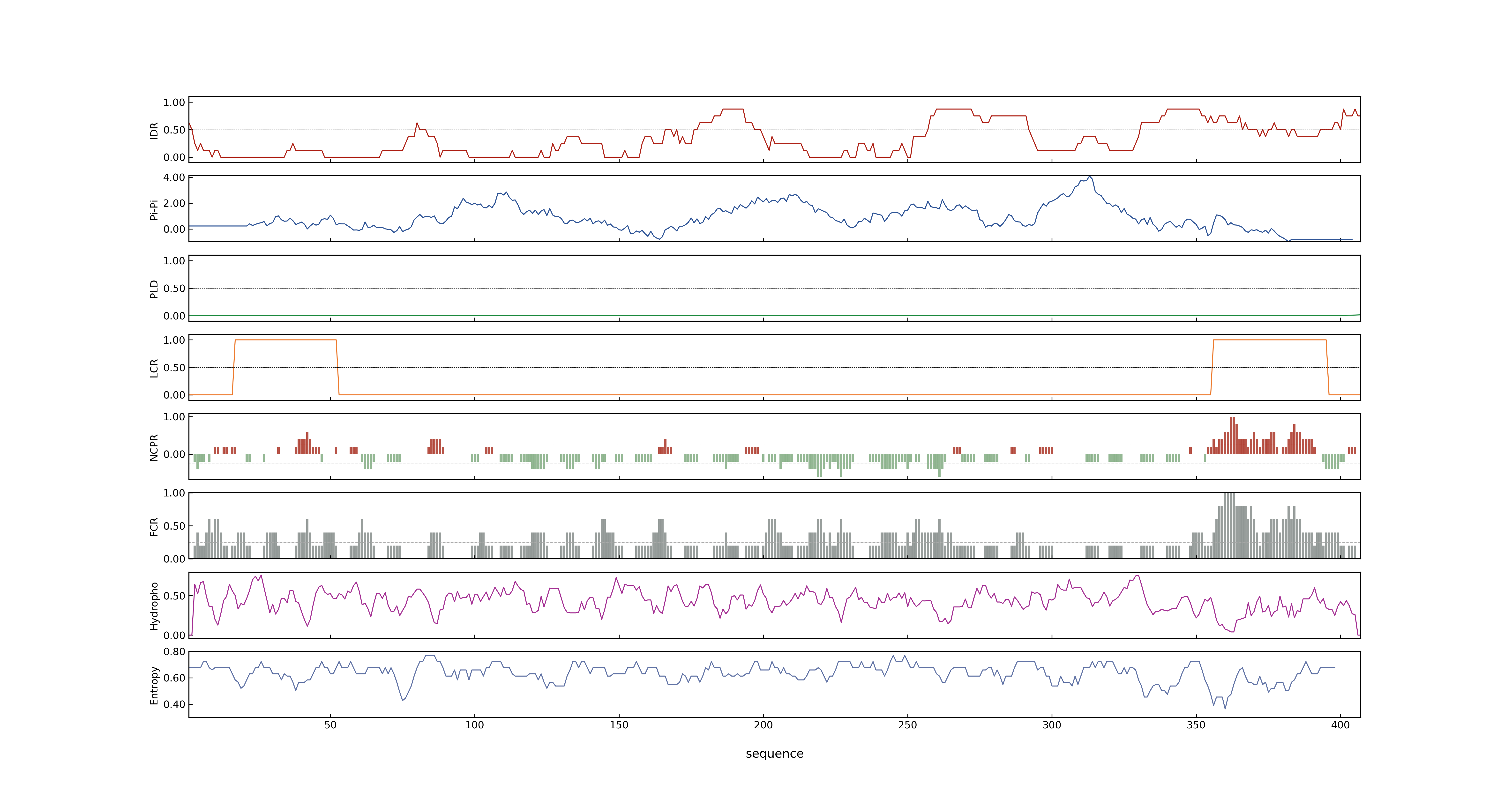
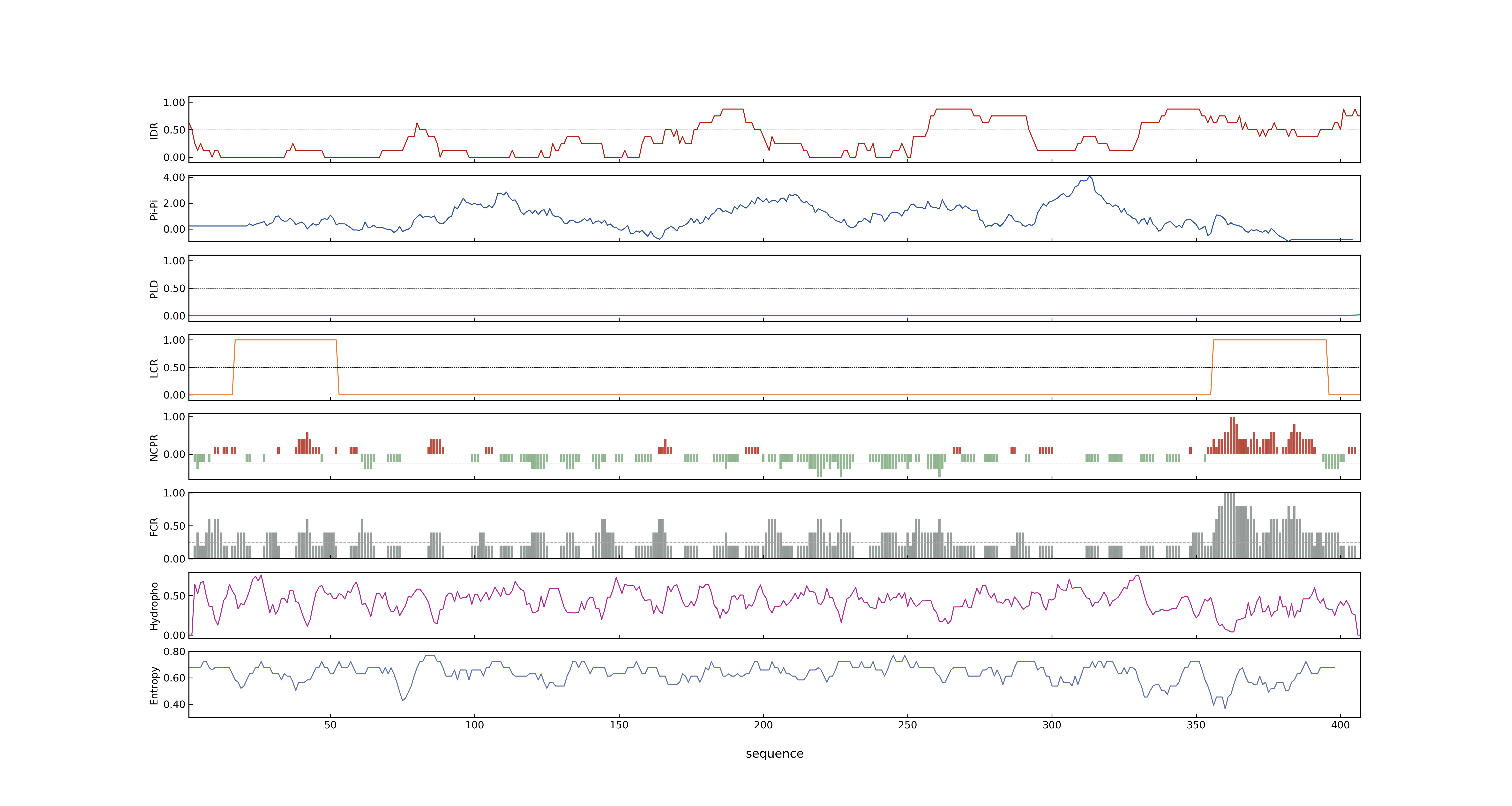
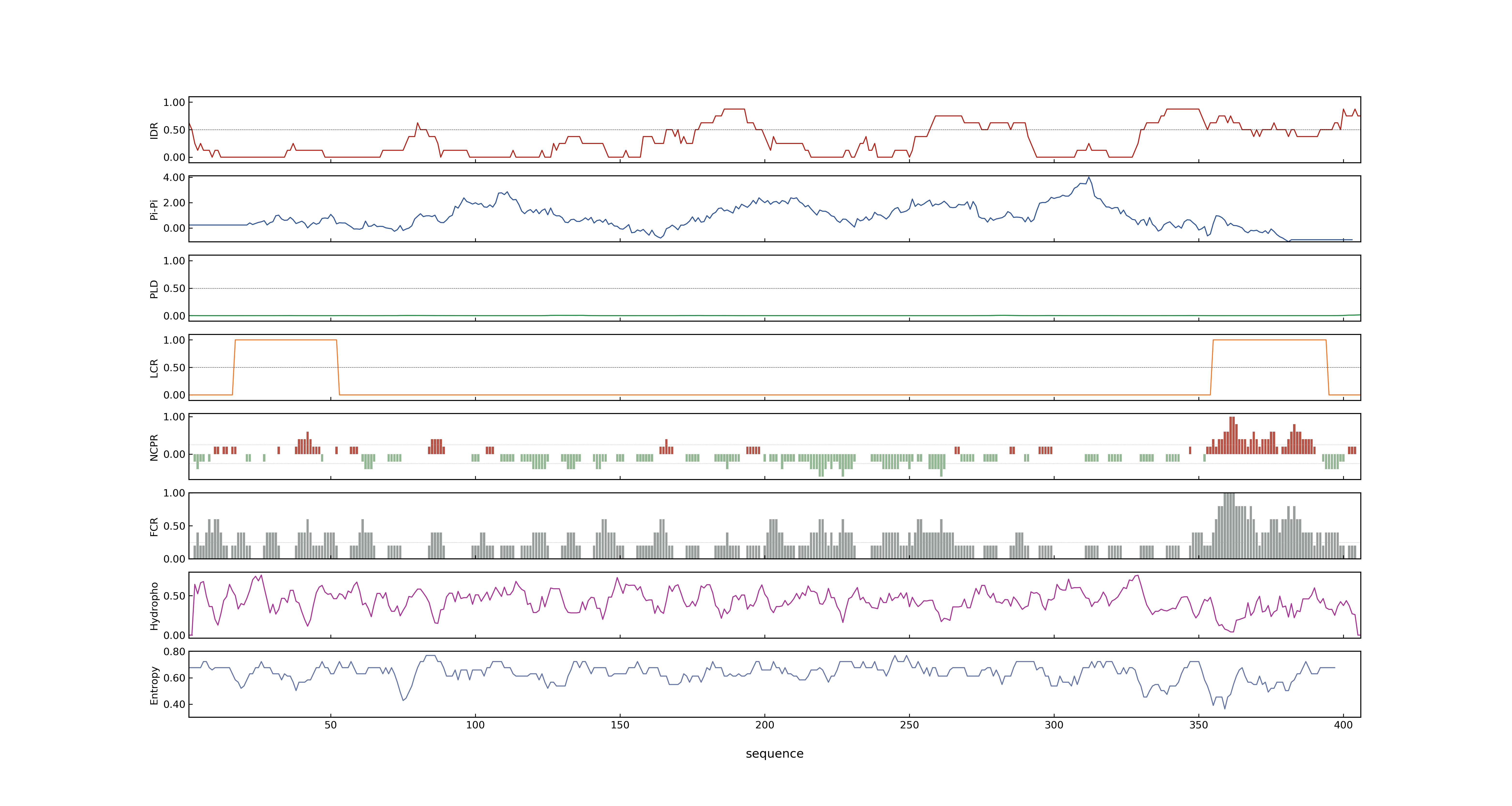
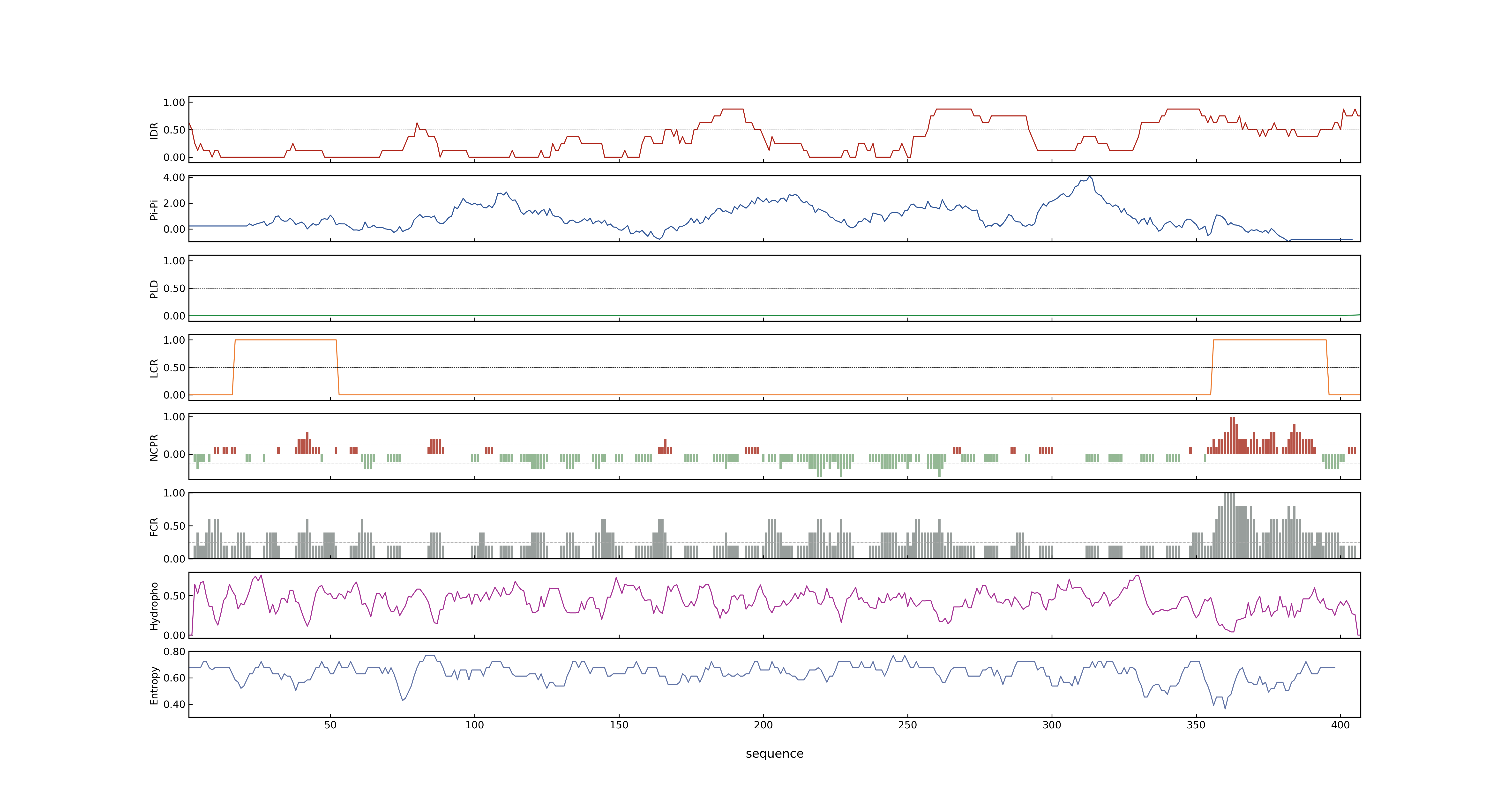
- PSP score
- LOC_Os02g49230.1: 0.5204
- LOC_Os02g49230.3: 0.5204
- LOC_Os02g49230.6: 0.5204
- LOC_Os02g49230.4: 0.5204
- LOC_Os02g49230.2: 0.7833
- LOC_Os02g49230.5: 0.5204
- PLAAC score
- LOC_Os02g49230.1: 0
- LOC_Os02g49230.3: 0
- LOC_Os02g49230.6: 0
- LOC_Os02g49230.4: 0
- LOC_Os02g49230.2: 0
- LOC_Os02g49230.5: 0
- pLDDT score
- 62.51
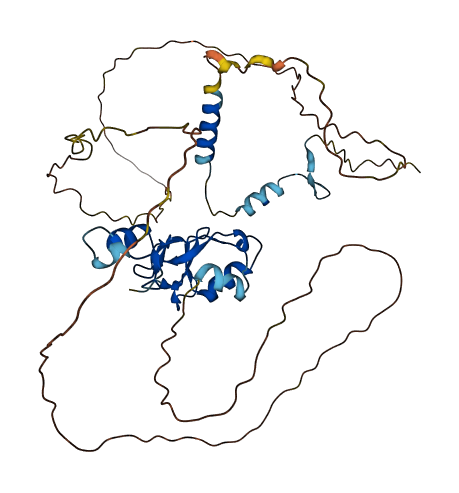
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os02g49230.1: 0.99970597
- LOC_Os02g49230.2: 0.99959117
- LOC_Os02g49230.3: 0.99970597
- LOC_Os02g49230.4: 0.99970597
- LOC_Os02g49230.5: 0.99970597
- LOC_Os02g49230.6: 0.99970597
- MolPhase Result
- Publication
- Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia, 2013, Proc Natl Acad Sci U S A.
- CONSTANS-Like 9 OsCOL9 Interacts with Receptor for Activated C-Kinase 1OsRACK1 to Regulate Blast Resistance through Salicylic Acid and Ethylene Signaling Pathways., 2016, PLoS One.
- CONSTANS-like 9 COL9 delays the flowering time in Oryza sativa by repressing the Ehd1 pathway., 2016, Biochem Biophys Res Commun.
- Genbank accession number
- Key message
- Our combined population genetics and network analyses suggest that DTH2 likely represents a target of human selection for adaptation to LD conditions during rice domestication and/or improvement, demonstrating an important role of minor-effect quantitative trait loci in crop adaptation and breeding
- We show that DTH2 encodes a CONSTANS-like protein that promotes heading by inducing the florigen genes Heading date 3a and RICE FLOWERING LOCUS T 1, and it acts independently of the known floral integrators Heading date 1 and Early heading date 1
- Moreover, association analysis and transgenic experiments identified two functional nucleotide polymorphisms in DTH2 that correlated with early heading and increased reproductive fitness under natural LD conditions in northern Asia
- In the present study, we investigated the functional roles of OsCOL9 in blast resistance
- Taken together, these results indicated that the COL protein OsCOL9 interacted with OsRACK1, and it enhanced the rice blast resistance through SA and ET signaling pathways
- OsCOL9 was a critical regulator of pathogen-related genes, especially PR1b, which were also activated by exogenous salicylic acid (SA) and 1-aminocyclopropane-1-carboxylicacid (ACC), the precursor of ethylene (ET)
- In a previous transcriptome analysis of early response genes in rice during Magnaporthe oryzae infection, we identified a CONSTANS-like (COL) gene OsCOL9
- Magnaporthe oryzae infection induced OsCOL9 expression, and transgenic OsCOL9 knock-out rice plants showed increased pathogen susceptibility
- OsCOL9 was found in the nucleus of rice cells, and it exerted transcriptional activation activities through its middle region (MR)
- Further analysis indicated that OsCOL9 over-expression increased the expressions of phytohormone biosynthetic genes, NPR1, WRKY45, OsACO1 and OsACS1, which were related to SA and ET biosynthesis
- Interestingly, we found that OsCOL9 physically interacted with the scaffold protein OsRACK1 through its CCT domain, and the OsRACK1 expression was induced in response to exogenous SA and ACC as well as M
- We have previously identified that the COL family member OsCOL9 can positively enhance the rice blast resistance
- In addition, OsCOL9 served as a potential yield gene, and its deficiency reduced the grain number of main panicle in plants
- OsCOL9 expression exhibited two types of circadian patterns under different daylight conditions, and it could delay the heading date by suppressing the Ehd1 photoperiodic flowering pathway
- Our data showed that overexpression of OsCOL9 delayed the flowering time under both short-day (SD) and long-day (LD) conditions, leading to suppressed expressions of EHd1, RFT and Hd3a at the mRNA Level
- Taken together, our results revealed that OsCOL9 could delay the flowering time in rice by repressing the Ehd1 pathway
- Connection
- DTH2~OsCOL9~OsCCT08, OsACO1, CONSTANS-Like 9 OsCOL9 Interacts with Receptor for Activated C-Kinase 1OsRACK1 to Regulate Blast Resistance through Salicylic Acid and Ethylene Signaling Pathways., Further analysis indicated that OsCOL9 over-expression increased the expressions of phytohormone biosynthetic genes, NPR1, WRKY45, OsACO1 and OsACS1, which were related to SA and ET biosynthesis
- DTH2~OsCOL9~OsCCT08, OsACS1, CONSTANS-Like 9 OsCOL9 Interacts with Receptor for Activated C-Kinase 1OsRACK1 to Regulate Blast Resistance through Salicylic Acid and Ethylene Signaling Pathways., Further analysis indicated that OsCOL9 over-expression increased the expressions of phytohormone biosynthetic genes, NPR1, WRKY45, OsACO1 and OsACS1, which were related to SA and ET biosynthesis
- DTH2~OsCOL9~OsCCT08, OsWRKY45~WRKY45, CONSTANS-Like 9 OsCOL9 Interacts with Receptor for Activated C-Kinase 1OsRACK1 to Regulate Blast Resistance through Salicylic Acid and Ethylene Signaling Pathways., Further analysis indicated that OsCOL9 over-expression increased the expressions of phytohormone biosynthetic genes, NPR1, WRKY45, OsACO1 and OsACS1, which were related to SA and ET biosynthesis
- DTH2~OsCOL9~OsCCT08, Ehd1, CONSTANS-like 9 COL9 delays the flowering time in Oryza sativa by repressing the Ehd1 pathway., Our data showed that overexpression of OsCOL9 delayed the flowering time under both short-day (SD) and long-day (LD) conditions, leading to suppressed expressions of EHd1, RFT and Hd3a at the mRNA Level
- DTH2~OsCOL9~OsCCT08, Ehd1, CONSTANS-like 9 COL9 delays the flowering time in Oryza sativa by repressing the Ehd1 pathway., OsCOL9 expression exhibited two types of circadian patterns under different daylight conditions, and it could delay the heading date by suppressing the Ehd1 photoperiodic flowering pathway
- DTH2~OsCOL9~OsCCT08, Ehd1, CONSTANS-like 9 COL9 delays the flowering time in Oryza sativa by repressing the Ehd1 pathway., Taken together, our results revealed that OsCOL9 could delay the flowering time in rice by repressing the Ehd1 pathway
- DTH2~OsCOL9~OsCCT08, Hd3a, CONSTANS-like 9 COL9 delays the flowering time in Oryza sativa by repressing the Ehd1 pathway., Our data showed that overexpression of OsCOL9 delayed the flowering time under both short-day (SD) and long-day (LD) conditions, leading to suppressed expressions of EHd1, RFT and Hd3a at the mRNA Level
Prev Next