- Information
- Symbol: ERS1,OsERS1
- MSU: LOC_Os03g49500
- RAPdb: Os03g0701700
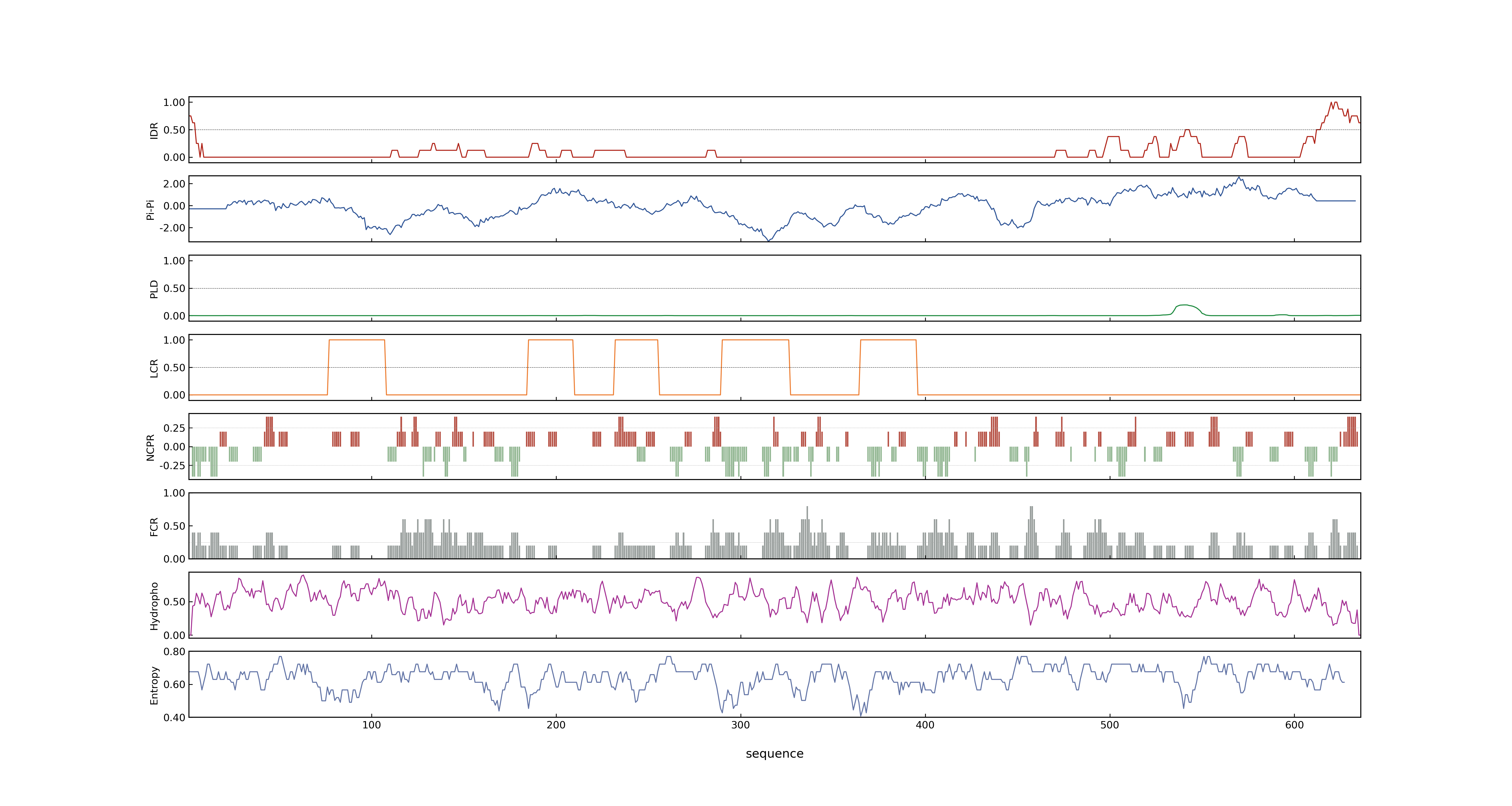
- PSP score
- LOC_Os03g49500.2: 0.0129
- LOC_Os03g49500.1: 0.0129
- PLAAC score
- LOC_Os03g49500.2: 0
- LOC_Os03g49500.1: 0
- pLDDT score
- NA
- MolPhase score
- LOC_Os03g49500.1: 0.98023122
- LOC_Os03g49500.2: 0.98023122
- MolPhase Result
- Publication
- Whole-genome analysis of Oryza sativa reveals similar architecture of two-component signaling machinery with Arabidopsis, 2006, Plant Physiol.
- Nomenclature for two-component signaling elements of rice, 2007, Plant Physiol.
- Differential expression of three genes encoding an ethylene receptor in rice during development, and in response to indole-3-acetic acid and silver ions, 2004, J Exp Bot.
- The Differentiated Localized Rice Ethylene Receptors OsERS1 and OsETR2 and their Potential Role during Submergence., 2017, Plant Signal Behav.
-
Genbank accession number
- Key message
- Northern analysis revealed that the level of OS-ETR2 mRNA was markedly elevated either by the exogenous application of IAA or by ethylene treatment in young etiolated rice seedlings, whereas the OS-ERS1 transcript level was only slightly induced under the same experimental conditions
- Pretreatment with silver prevented IAA-induced and ethylene-induced accumulation of both mRNAs (OS-ERS1 and OS-ETR2)
- Five ethylene receptor genes, OS-ERS1, OS-ERS2, OS-ETR2, OS-ETR3, and OS-ETR4 were isolated and characterized from rice
- Deduced amino acid sequences of OS-ERS1, OS-ERS2, OS-ETR2, OS-ETR3, and OS-ETR4 showed that they exhibited significant homology to the prokaryotic two-component signal transducer and a wide range of ethylene receptors in a variety of plant species
- Analysis of the expression of the three ethylene receptor genes in different tissues of rice has unravelled their corresponding tissue-specificity in which OS-ERS1 was constitutively expressed in considerable amounts in all tissues studied, while OS-ERS2 and OS-ETR2 exhibited differential expression patterns in different tissues of rice
- Our results and others support the notion that OsERS1 and OsETR2 could have different roles during rice plant submergence
- The Differentiated Localized Rice Ethylene Receptors OsERS1 and OsETR2 and their Potential Role during Submergence.
- OsERS1 and OsETR2 are major ethylene receptors in rice that have been reported to have different regulatory functions
- Base on the results, we suggested that OsERS1 could be localized to plasma membranes, whereas OsETR2 could be localized to the endoplasmic reticulum
- Connection
- ERS1~OsERS1, ERS2, Differential expression of three genes encoding an ethylene receptor in rice during development, and in response to indole-3-acetic acid and silver ions, Five ethylene receptor genes, OS-ERS1, OS-ERS2, OS-ETR2, OS-ETR3, and OS-ETR4 were isolated and characterized from rice
- ERS1~OsERS1, ERS2, Differential expression of three genes encoding an ethylene receptor in rice during development, and in response to indole-3-acetic acid and silver ions, The genomic structure of OS-ERS1 and OS-ERS2 revealed that the introns within the coding sequences occurred in conserved positions to those of At-ETR1 and At-ERS1, whereas each of the OS-ETR2, OS-ETR3, and OS-ETR4 genes contained 1 intron within its coding region located at a position equivalent to those of At-ERS2, At-ETR2, and At-EIN4
- ERS1~OsERS1, ERS2, Differential expression of three genes encoding an ethylene receptor in rice during development, and in response to indole-3-acetic acid and silver ions, Deduced amino acid sequences of OS-ERS1, OS-ERS2, OS-ETR2, OS-ETR3, and OS-ETR4 showed that they exhibited significant homology to the prokaryotic two-component signal transducer and a wide range of ethylene receptors in a variety of plant species
- ERS1~OsERS1, ERS2, Differential expression of three genes encoding an ethylene receptor in rice during development, and in response to indole-3-acetic acid and silver ions, Analysis of the expression of the three ethylene receptor genes in different tissues of rice has unravelled their corresponding tissue-specificity in which OS-ERS1 was constitutively expressed in considerable amounts in all tissues studied, while OS-ERS2 and OS-ETR2 exhibited differential expression patterns in different tissues of rice
- ERS1~OsERS1, ETR3, Differential expression of three genes encoding an ethylene receptor in rice during development, and in response to indole-3-acetic acid and silver ions, Five ethylene receptor genes, OS-ERS1, OS-ERS2, OS-ETR2, OS-ETR3, and OS-ETR4 were isolated and characterized from rice
- ERS1~OsERS1, ETR3, Differential expression of three genes encoding an ethylene receptor in rice during development, and in response to indole-3-acetic acid and silver ions, The genomic structure of OS-ERS1 and OS-ERS2 revealed that the introns within the coding sequences occurred in conserved positions to those of At-ETR1 and At-ERS1, whereas each of the OS-ETR2, OS-ETR3, and OS-ETR4 genes contained 1 intron within its coding region located at a position equivalent to those of At-ERS2, At-ETR2, and At-EIN4
- ERS1~OsERS1, ETR3, Differential expression of three genes encoding an ethylene receptor in rice during development, and in response to indole-3-acetic acid and silver ions, Deduced amino acid sequences of OS-ERS1, OS-ERS2, OS-ETR2, OS-ETR3, and OS-ETR4 showed that they exhibited significant homology to the prokaryotic two-component signal transducer and a wide range of ethylene receptors in a variety of plant species
- ERS1~OsERS1, ETR4, Differential expression of three genes encoding an ethylene receptor in rice during development, and in response to indole-3-acetic acid and silver ions, Five ethylene receptor genes, OS-ERS1, OS-ERS2, OS-ETR2, OS-ETR3, and OS-ETR4 were isolated and characterized from rice
- ERS1~OsERS1, ETR4, Differential expression of three genes encoding an ethylene receptor in rice during development, and in response to indole-3-acetic acid and silver ions, The genomic structure of OS-ERS1 and OS-ERS2 revealed that the introns within the coding sequences occurred in conserved positions to those of At-ETR1 and At-ERS1, whereas each of the OS-ETR2, OS-ETR3, and OS-ETR4 genes contained 1 intron within its coding region located at a position equivalent to those of At-ERS2, At-ETR2, and At-EIN4
- ERS1~OsERS1, ETR4, Differential expression of three genes encoding an ethylene receptor in rice during development, and in response to indole-3-acetic acid and silver ions, Deduced amino acid sequences of OS-ERS1, OS-ERS2, OS-ETR2, OS-ETR3, and OS-ETR4 showed that they exhibited significant homology to the prokaryotic two-component signal transducer and a wide range of ethylene receptors in a variety of plant species
Prev Next