- Information
- Symbol: Ghd7
- MSU: LOC_Os07g15770
- RAPdb: Os07g0261200
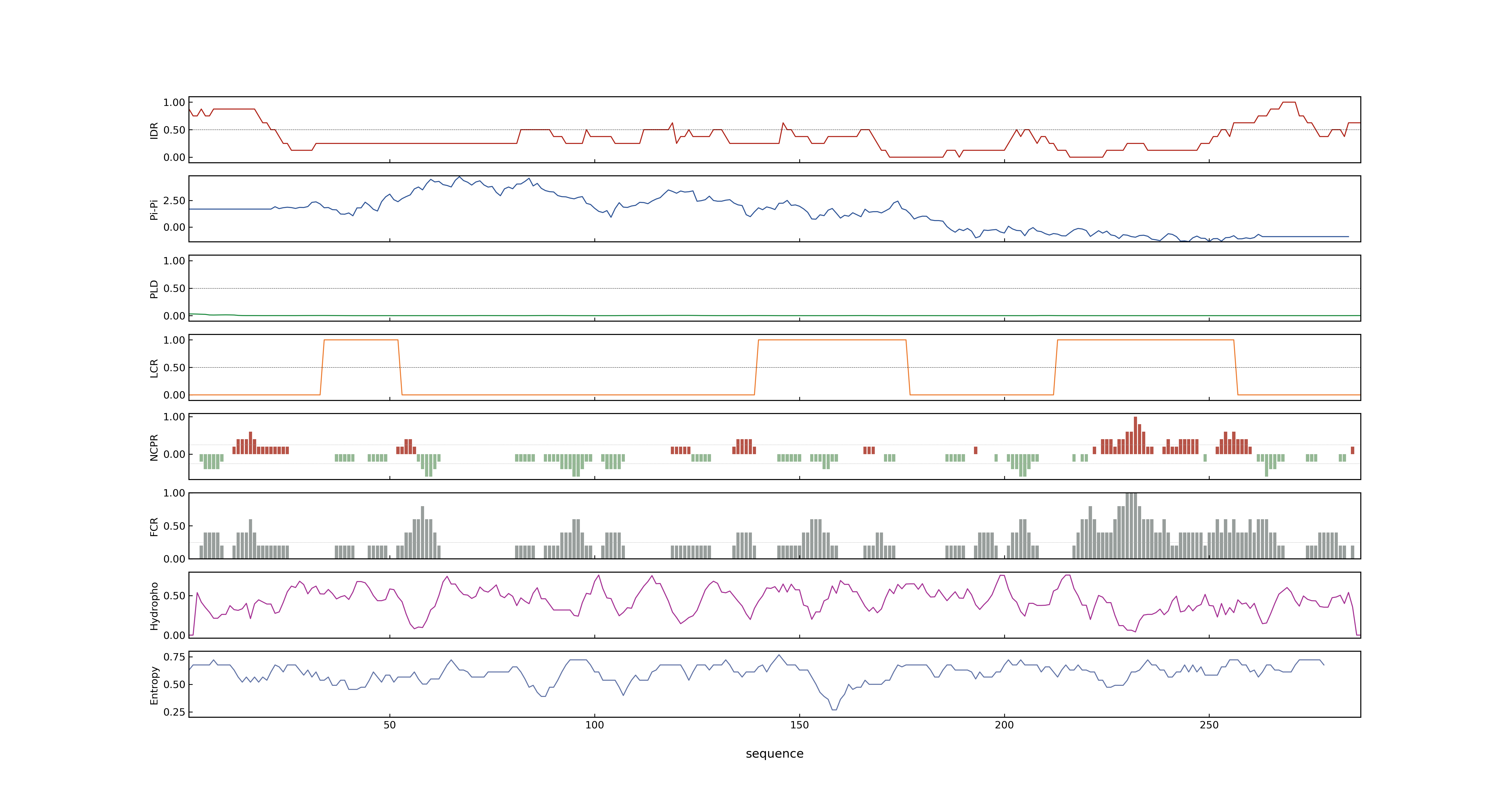
- PSP score
- LOC_Os07g15770.1: 0.5815
- PLAAC score
- LOC_Os07g15770.1: 0
(Zheng et al., New Phytol, 2019, 224:306-320)
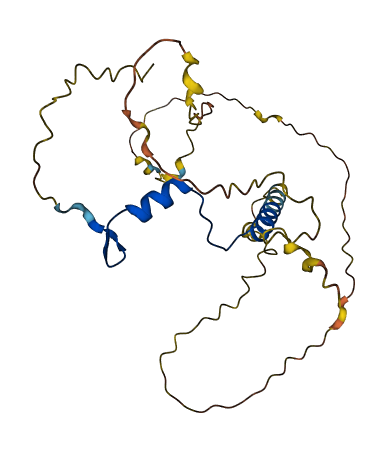
- pLDDT score
- 61.33
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os07g15770.1: 0.99984991
- MolPhase Result
- Publication
- Genetic interactions involved in the inhibition of heading by heading date QTL, Hd2 in rice under long-day conditions, 2011, Theor Appl Genet.
- Roles of the Hd5 gene controlling heading date for adaptation to the northern limits of rice cultivation, 2013, Theor Appl Genet.
- Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice, 2011, Plant Physiol.
- A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice, 2009, Development.
- OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, 2009, Plant Cell Environ.
- Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering, 2011, Plant J.
- Evolution and association analysis of Ghd7 in rice, 2012, PLoS One.
- OsELF3-1, an ortholog of Arabidopsis early flowering 3, regulates rice circadian rhythm and photoperiodic flowering, 2012, PLoS One.
- Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes, 2013, Mol Plant.
- Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice, 2008, Nat Genet.
- A pair of floral regulators sets critical day length for Hd3a florigen expression in rice, 2010, Nat Genet.
- Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering, 2012, Plant Cell Physiol.
- DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously, 2010, Plant Physiol.
- Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date3, 2014, Plant Physiol.
- Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response, 2013, Plant J.
- Ghd7 is a central regulator for growth, development, adaptation and responses to biotic and abiotic stresses, 2014, Plant physiology.
- Genetic architecture of natural variation in rice chlorophyll content revealed by genome wide association study., 2015, Mol Plant.
- Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice., 2015, New Phytol.
- Hd1,a CONSTANS Orthlog in Rice, Functions as an Ehd1 Repressor Through Interaction with Monocot-Specific CCT-Domain Protein Ghd7., 2016, Plant J.
- Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions., 2017, Sci Rep.
- Genbank accession number
- Key message
- Here we show that the quantitative trait locus (QTL) Ghd7, isolated from an elite rice hybrid and encoding a CCT domain protein, has major effects on an array of traits in rice, including number of grains per panicle, plant height and heading date
- Enhanced expression of Ghd7 under long-day conditions delays heading and increases plant height and panicle size
- Moreover, OsELF3-1 suppresses a flowering repressor grain number, plant height and heading date 7 (Ghd7) to indirectly accelerate flowering under long-day (LD) conditions
- Ehd1 and Hd3a can also be down-regulated by the photoperiodic flowering genes Ghd7 and Hd1 (a rice ortholog of CONSTANS)
- By using near-isogenic lines with functional or deficient alleles of several rice flowering-time genes, we observed significant digenetic interactions between Hd16 and four other flowering-time genes (Ghd7, Hd1, DTH8 and Hd2)
- These results demonstrate that Hd16 acts as an inhibitor in the rice flowering pathway by enhancing the photoperiod response as a result of the phosphorylation of Ghd7
- Our results demonstrate that natural variation in Hd17 may change the transcription level of a flowering repressor, Grain number, plant height and heading date 7 (Ghd7), suggesting that Hd17 is part of rice’s photoperiodic flowering pathway
- On the other hand, under long-day (LD) conditions, flowering is delayed by the repressive function of Hd1 on FT-like genes and by downregulation of Ehd1 by the flowering repressor Ghd7 - a unique pathway in rice
- Furthermore, Ehd3 ghd7 plants flowered earlier and show higher Ehd1 transcript levels than ehd3 ghd7 plants, suggesting a Ghd7-independent role of Ehd3 in the upregulation of Ehd1
- We propose that both positive (OsMADS50 and Ehd1) and negative (Hd1, phyB and Ghd7) regulators of RFT1 form a gene network that regulates LD flowering in rice
- Ghd7 (Grain number, plant height and heading date 7) was acutely induced when phytochrome signals coincided with a photosensitive phase set differently by distinct photoperiods and this induction repressed Ehd1 the next morning
- However, the Grain number, plant height, and heading date7 (Ghd7) pathway was altered in ostrx1
- Increased transcription of Ghd7 under LD conditions and reduced transcription of downstream Ehd1 and FT-like genes in the ehd3 mutants suggested that Ehd3 normally functions as an LD downregulator of Ghd7 in floral induction
- rufipogon) was used to analyze the evolution and association of Ghd7 with plant height, heading date, and yield
- Meanwhile, the transcription of DTH8 has been proved to be independent of Ghd7 and Hd1, and the natural mutation of this gene caused weak photoperiod sensitivity and shorter plant height
- Analyses using a rice phytochrome chromophore-deficient mutant, photoperiod sensitivity5, have so far revealed that within this network, phytochromes are required for expression of Grain number, plant height and heading date7 (Ghd7), a floral repressor gene in rice
- Similarly, in the homozygous Koshihikari genetic background at Ghd7, the difference in heading date caused by different alleles at Hd2 was smaller than in plants homozygous for the Hayamasari Ghd7 allele
- In addition, QTLs near Hd2, Hd16, and Ghd7, which are involved in inhibition of heading under long-day conditions, function in the same pathway that controls heading date
- SNP changes between haplotypes indicated that Ghd7 evolved from two distinct ancestral gene pools, and independent domestication processes were detected in indica and japonica varietals respectively
- Thus, two distinct gating mechanisms–of the floral promoter Ehd1 and the floral repressor Ghd7–could enable manipulation of slight differences in day length to control Hd3a transcription with a critical day-length threshold
- Transcript levels of OsGI, phytochrome genes, and Early heading date3 (Ehd3), which function upstream of Ghd7, were unchanged in the mutant
- Moreover, phyB and phyA can affect Ghd7 activity and Early heading date1 (a floral inducer) activity in the network, respectively
- These associations provide the potential for flexibility of Ghd7 application in rice breeding programs
- Notably, the japonica varieties harboring nonfunctional alleles of both Ghd7/Hd4 and PRR37/Hd2 flower extremely early under natural long-day conditions, and are adapted to the northernmost regions of rice cultivation, up to 53 degrees N latitude
- Genetic analysis revealed that the effects of PRR37 and Ghd7 alleles on heading date are additive, and PRR37 down-regulates Hd3a expression to suppress flowering under long-day conditions
- Thus, Ghd7 has played crucial roles for increasing productivity and adaptability of rice globally
- Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice
- The enhanced expression of Ghd7 decreased chlorophyll content, mainly through down-regulating the expression of genes involved in the biosynthesis of chlorophyll and chloroplast
- We employed a map-based cloning approach to isolate a heading date gene, which coordinated the interaction between Ghd7 and Ghd8 to greatly delay rice heading
- Map-based cloning demonstrated that the gene largely affecting the interaction between Ghd7 and Ghd8 was Hd1
- Here, we report biological interactions between Ghd7 and Hd1, which together repress Early heading date 1 (Ehd1), a key floral inducer under non-inductive long-day (LD) conditions
- These findings imply that Hd1, an evolutionally conserved transcriptional activator, can function as a strong transcriptional repressor within a monocot-specific flowering-time pathway through with Ghd7
- These findings suggest that Hd1 alone essentially acts as a promoter of heading date, and the protein interaction between Ghd7 and Hd1 determines photoperiod sensitivity and integrated Hd1-mediated and Ehd1-mediated flowering pathways in rice
- Connection
- Ghd7, Hd5~DTH8~Ghd8~OsHAP3H~LHD1~EF8~CAR8~OsNF-YB11, Genetic interactions involved in the inhibition of heading by heading date QTL, Hd2 in rice under long-day conditions, Combinations of several QTLs near Hd1, Hd2, Ghd7, Hd5, and Hd16 were detected under these four conditions
- Ghd7, Hd1, Genetic interactions involved in the inhibition of heading by heading date QTL, Hd2 in rice under long-day conditions, Combinations of several QTLs near Hd1, Hd2, Ghd7, Hd5, and Hd16 were detected under these four conditions
- CKI~EL1~Hd16, Ghd7, Genetic interactions involved in the inhibition of heading by heading date QTL, Hd2 in rice under long-day conditions, Combinations of several QTLs near Hd1, Hd2, Ghd7, Hd5, and Hd16 were detected under these four conditions
- CKI~EL1~Hd16, Ghd7, Genetic interactions involved in the inhibition of heading by heading date QTL, Hd2 in rice under long-day conditions, Analysis of advanced backcross progenies revealed genetic interactions between Hd2 and Hd16 and between Hd2 and Ghd7
- CKI~EL1~Hd16, Ghd7, Genetic interactions involved in the inhibition of heading by heading date QTL, Hd2 in rice under long-day conditions, In addition, QTLs near Hd2, Hd16, and Ghd7, which are involved in inhibition of heading under long-day conditions, function in the same pathway that controls heading date
- Ghd7, Hd5~DTH8~Ghd8~OsHAP3H~LHD1~EF8~CAR8~OsNF-YB11, Roles of the Hd5 gene controlling heading date for adaptation to the northern limits of rice cultivation, The loss-of-function Hd5 was then introduced into the rice variety Fanny from France and contributed to its extremely early heading under the presence of functional Ghd7
- Ghd7, PHYA~OsPhyA, Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice, Our results show that either phyA alone or a genetic combination of phyB and phyC can induce Ghd7 mRNA, whereas phyB alone causes some reduction in levels of Ghd7 mRNA
- Ghd7, PHYA~OsPhyA, Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice, Moreover, phyB and phyA can affect Ghd7 activity and Early heading date1 (a floral inducer) activity in the network, respectively
- Ghd7, PHYB~OsphyB, Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice, Our results show that either phyA alone or a genetic combination of phyB and phyC can induce Ghd7 mRNA, whereas phyB alone causes some reduction in levels of Ghd7 mRNA
- Ghd7, PHYB~OsphyB, Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice, Moreover, phyB and phyA can affect Ghd7 activity and Early heading date1 (a floral inducer) activity in the network, respectively
- Ghd7, PHYC, Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice, Our results show that either phyA alone or a genetic combination of phyB and phyC can induce Ghd7 mRNA, whereas phyB alone causes some reduction in levels of Ghd7 mRNA
- Ghd7, Hd1, A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice, We propose that both positive (OsMADS50 and Ehd1) and negative (Hd1, phyB and Ghd7) regulators of RFT1 form a gene network that regulates LD flowering in rice
- Ghd7, PHYB~OsphyB, A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice, We propose that both positive (OsMADS50 and Ehd1) and negative (Hd1, phyB and Ghd7) regulators of RFT1 form a gene network that regulates LD flowering in rice
- Ehd1, Ghd7, A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice, We propose that both positive (OsMADS50 and Ehd1) and negative (Hd1, phyB and Ghd7) regulators of RFT1 form a gene network that regulates LD flowering in rice
- Ghd7, RFT1, A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice, We propose that both positive (OsMADS50 and Ehd1) and negative (Hd1, phyB and Ghd7) regulators of RFT1 form a gene network that regulates LD flowering in rice
- Ghd7, OsMADS50~OsSOC1~DTH3, A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice, We propose that both positive (OsMADS50 and Ehd1) and negative (Hd1, phyB and Ghd7) regulators of RFT1 form a gene network that regulates LD flowering in rice
- Ghd7, Hd6~CK2, OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, On the other hand, mRNA levels of OsGI, Hd1, OsId1, OsDof12, Ghd7, Hd6 and SE5 were unchanged
- Ghd7, OsDof12~OsCDF1, OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, On the other hand, mRNA levels of OsGI, Hd1, OsId1, OsDof12, Ghd7, Hd6 and SE5 were unchanged
- Ghd7, OsGI, OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, On the other hand, mRNA levels of OsGI, Hd1, OsId1, OsDof12, Ghd7, Hd6 and SE5 were unchanged
- Ghd7, Hd1, OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, On the other hand, mRNA levels of OsGI, Hd1, OsId1, OsDof12, Ghd7, Hd6 and SE5 were unchanged
- Ehd1, Ghd7, Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering, On the other hand, under long-day (LD) conditions, flowering is delayed by the repressive function of Hd1 on FT-like genes and by downregulation of Ehd1 by the flowering repressor Ghd7 - a unique pathway in rice
- Ehd1, Ghd7, Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering, Increased transcription of Ghd7 under LD conditions and reduced transcription of downstream Ehd1 and FT-like genes in the ehd3 mutants suggested that Ehd3 normally functions as an LD downregulator of Ghd7 in floral induction
- Ehd1, Ghd7, Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering, Furthermore, Ehd3 ghd7 plants flowered earlier and show higher Ehd1 transcript levels than ehd3 ghd7 plants, suggesting a Ghd7-independent role of Ehd3 in the upregulation of Ehd1
- Ghd7, Hd1, Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering, On the other hand, under long-day (LD) conditions, flowering is delayed by the repressive function of Hd1 on FT-like genes and by downregulation of Ehd1 by the flowering repressor Ghd7 - a unique pathway in rice
- Ehd3, Ghd7, Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering, Increased transcription of Ghd7 under LD conditions and reduced transcription of downstream Ehd1 and FT-like genes in the ehd3 mutants suggested that Ehd3 normally functions as an LD downregulator of Ghd7 in floral induction
- Ehd3, Ghd7, Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering, Furthermore, Ehd3 ghd7 plants flowered earlier and show higher Ehd1 transcript levels than ehd3 ghd7 plants, suggesting a Ghd7-independent role of Ehd3 in the upregulation of Ehd1
- Ghd7, Hd17~Ef7~OsELF3-1~OsELF3.1, OsELF3-1, an ortholog of Arabidopsis early flowering 3, regulates rice circadian rhythm and photoperiodic flowering, Moreover, OsELF3-1 suppresses a flowering repressor grain number, plant height and heading date 7 (Ghd7) to indirectly accelerate flowering under long-day (LD) conditions
- Ehd1, Ghd7, A pair of floral regulators sets critical day length for Hd3a florigen expression in rice, Ghd7 (Grain number, plant height and heading date 7) was acutely induced when phytochrome signals coincided with a photosensitive phase set differently by distinct photoperiods and this induction repressed Ehd1 the next morning
- Ehd1, Ghd7, A pair of floral regulators sets critical day length for Hd3a florigen expression in rice, Thus, two distinct gating mechanisms–of the floral promoter Ehd1 and the floral repressor Ghd7–could enable manipulation of slight differences in day length to control Hd3a transcription with a critical day-length threshold
- Ghd7, Hd3a, A pair of floral regulators sets critical day length for Hd3a florigen expression in rice, Thus, two distinct gating mechanisms–of the floral promoter Ehd1 and the floral repressor Ghd7–could enable manipulation of slight differences in day length to control Hd3a transcription with a critical day-length threshold
- Ghd7, Hd17~Ef7~OsELF3-1~OsELF3.1, Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering, Our results demonstrate that natural variation in Hd17 may change the transcription level of a flowering repressor, Grain number, plant height and heading date 7 (Ghd7), suggesting that Hd17 is part of rice’s photoperiodic flowering pathway
- Ghd7, Hd5~DTH8~Ghd8~OsHAP3H~LHD1~EF8~CAR8~OsNF-YB11, DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously, Meanwhile, the transcription of DTH8 has been proved to be independent of Ghd7 and Hd1, and the natural mutation of this gene caused weak photoperiod sensitivity and shorter plant height
- Ehd1, Ghd7, DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously, Ehd1 and Hd3a can also be down-regulated by the photoperiodic flowering genes Ghd7 and Hd1 (a rice ortholog of CONSTANS)
- Ghd7, Hd1, DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously, Ehd1 and Hd3a can also be down-regulated by the photoperiodic flowering genes Ghd7 and Hd1 (a rice ortholog of CONSTANS)
- Ghd7, Hd1, DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously, Meanwhile, the transcription of DTH8 has been proved to be independent of Ghd7 and Hd1, and the natural mutation of this gene caused weak photoperiod sensitivity and shorter plant height
- Ghd7, Hd3a, DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously, Ehd1 and Hd3a can also be down-regulated by the photoperiodic flowering genes Ghd7 and Hd1 (a rice ortholog of CONSTANS)
- Ehd3, Ghd7, Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date3, Transcript levels of OsGI, phytochrome genes, and Early heading date3 (Ehd3), which function upstream of Ghd7, were unchanged in the mutant
- Ghd7, OsGI, Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date3, Transcript levels of OsGI, phytochrome genes, and Early heading date3 (Ehd3), which function upstream of Ghd7, were unchanged in the mutant
- Ghd7, OsTrx1~SDG723, Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date3, However, the Grain number, plant height, and heading date7 (Ghd7) pathway was altered in ostrx1
- Ghd7, OsTrx1~SDG723, Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date3, Because Trx group proteins form a complex with other proteins to modify the chromatin structure of target genes, we investigated whether OsTrx1 interacts with a previously identified protein that functions upstream of Ghd7
- Ghd7, Hd5~DTH8~Ghd8~OsHAP3H~LHD1~EF8~CAR8~OsNF-YB11, Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response, By using near-isogenic lines with functional or deficient alleles of several rice flowering-time genes, we observed significant digenetic interactions between Hd16 and four other flowering-time genes (Ghd7, Hd1, DTH8 and Hd2)
- CKI~EL1~Hd16, Ghd7, Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response, By using near-isogenic lines with functional or deficient alleles of several rice flowering-time genes, we observed significant digenetic interactions between Hd16 and four other flowering-time genes (Ghd7, Hd1, DTH8 and Hd2)
- CKI~EL1~Hd16, Ghd7, Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response, Biochemical characterization indicated that the functional Hd16 recombinant protein specifically phosphorylated Ghd7
- CKI~EL1~Hd16, Ghd7, Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response, These results demonstrate that Hd16 acts as an inhibitor in the rice flowering pathway by enhancing the photoperiod response as a result of the phosphorylation of Ghd7
- Ghd7, Hd1, Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response, By using near-isogenic lines with functional or deficient alleles of several rice flowering-time genes, we observed significant digenetic interactions between Hd16 and four other flowering-time genes (Ghd7, Hd1, DTH8 and Hd2)
- Ghd7, Ghd7.1~Hd2~OsPRR37~DTH7, Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice., Further, we find that haplotype combinations of DTH7 with Grain number, plant height, and heading date 7 (Ghd7) and DTH8 correlate well with the heading date and grain yield of rice under different photoperiod conditions
- CKI~EL1~Hd16, Ghd7, Casein Kinases I and 2ÃÂÃÂÃÂÃÂÃÂÃÂÃÂÃÂÃÂÃÂÃÂÃÂÃÂÃÂÃÂñ Phosphorylate Oryza Sativa Pseudo-Response Regulator 37 OsPRR37 in Photoperiodic Flowering in Rice, In floral induction, Hd16 acts upstream of Ghd7 and CKI interacts with and phosphorylates Ghd7
- FKF1~OsFKF1, Ghd7, Rice FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 OsFKF1 promotes flowering independent of photoperiod., Moreover, OsFKF1 can upregulate Ehd1 expression under blue light treatment, without affecting the expression of Ehd2 and Ghd7
- FKF1~OsFKF1, Ghd7, Rice FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 OsFKF1 promotes flowering independent of photoperiod., In contrast to the LD-specific floral activator Arabidopsis FKF1, OsFKF1 likely acts as an autonomous floral activator because it promotes flowering independent of photoperiod, probably via its distinct roles in controlling expression of rice-specific genes including Ehd2, Ghd7, and Ehd1
- Ghd7, Hd1, Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice., Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice.
- Ghd7, Hd1, Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice., Map-based cloning demonstrated that the gene largely affecting the interaction between Ghd7 and Ghd8 was Hd1
- Ghd7, Hd1, Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice., Natural variation analysis showed that a combination of loss-of-function alleles of Ghd7, Ghd8 and Hd1 contributes to the expansion of rice cultivars to higher latitudes; by contrast, a combination of pre-existing strong alleles of Ghd7, Ghd8 and functional Hd1 (referred as SSF) is exclusively found where ancestral Asian cultivars originated
- Ghd7, Hd1, Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice., Our results indicate that the combinations of Ghd7, Ghd8 and Hd1 largely define the ecogeographical adaptation and yield potential in rice cultivars
- Ghd7, Hd5~DTH8~Ghd8~OsHAP3H~LHD1~EF8~CAR8~OsNF-YB11, Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice., Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice.
- Ghd7, Hd5~DTH8~Ghd8~OsHAP3H~LHD1~EF8~CAR8~OsNF-YB11, Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice., We employed a map-based cloning approach to isolate a heading date gene, which coordinated the interaction between Ghd7 and Ghd8 to greatly delay rice heading
- Ghd7, Hd5~DTH8~Ghd8~OsHAP3H~LHD1~EF8~CAR8~OsNF-YB11, Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice., Map-based cloning demonstrated that the gene largely affecting the interaction between Ghd7 and Ghd8 was Hd1
- Ghd7, Hd5~DTH8~Ghd8~OsHAP3H~LHD1~EF8~CAR8~OsNF-YB11, Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice., Natural variation analysis showed that a combination of loss-of-function alleles of Ghd7, Ghd8 and Hd1 contributes to the expansion of rice cultivars to higher latitudes; by contrast, a combination of pre-existing strong alleles of Ghd7, Ghd8 and functional Hd1 (referred as SSF) is exclusively found where ancestral Asian cultivars originated
- Ghd7, Hd5~DTH8~Ghd8~OsHAP3H~LHD1~EF8~CAR8~OsNF-YB11, Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice., Our results indicate that the combinations of Ghd7, Ghd8 and Hd1 largely define the ecogeographical adaptation and yield potential in rice cultivars
- Ghd7, Ghd7.1~Hd2~OsPRR37~DTH7, OsPRR37 and Ghd7 are the major genes for general combining ability of DTH, PH and SPP in rice., OsPRR37 and Ghd7 are the major genes for general combining ability of DTH, PH and SPP in rice.
- Ghd7, Ghd7.1~Hd2~OsPRR37~DTH7, OsPRR37 and Ghd7 are the major genes for general combining ability of DTH, PH and SPP in rice., Both GCA1 and GCA2 were loss-of-function gene in low-GCA parent and gain-of-function gene in high-GCA parent, encoding the putative Pseudo-Response Regulators, OsPRR37 and Ghd7, respectively
- Ghd7, Ghd7.1~Hd2~OsPRR37~DTH7, OsPRR37 and Ghd7 are the major genes for general combining ability of DTH, PH and SPP in rice., Our results demonstrate that two GCA loci associate with OsPRR37 and Ghd7 and reveal that the genes responsible for important agronomic traits could simultaneously account for GCA effects
- Ghd7, OsCOL10, OsCOL10, a CONSTANS-like gene, functions as a flowering-time repressor downstream of Ghd7 in rice., OsCOL10, a CONSTANS-like gene, functions as a flowering-time repressor downstream of Ghd7 in rice.
- Ghd7, OsCOL10, OsCOL10, a CONSTANS-like gene, functions as a flowering-time repressor downstream of Ghd7 in rice., Transcripts of OsCOL10 are more abundant in the plants carrying a functional Ghd7 allele or over-expressing Ghd7 than in the Ghd7-deficient ones, thus placing OsCOL10 downstream of Ghd7
- Ghd7, OsCOL10, OsCOL10, a CONSTANS-like gene, functions as a flowering-time repressor downstream of Ghd7 in rice., Together, we conclude that OsCOL10 functions as a flowering-time repressor that links between Ghd7 and Ehd1 in rice
- Ehd1, Ghd7, Hd1,a CONSTANS Orthlog in Rice, Functions as an Ehd1 Repressor Through Interaction with Monocot-Specific CCT-Domain Protein Ghd7., Hd1,a CONSTANS Orthlog in Rice, Functions as an Ehd1 Repressor Through Interaction with Monocot-Specific CCT-Domain Protein Ghd7.
- Ehd1, Ghd7, Hd1,a CONSTANS Orthlog in Rice, Functions as an Ehd1 Repressor Through Interaction with Monocot-Specific CCT-Domain Protein Ghd7., Here, we report biological interactions between Ghd7 and Hd1, which together repress Early heading date 1 (Ehd1), a key floral inducer under non-inductive long-day (LD) conditions
- Ghd7, Hd1, Hd1,a CONSTANS Orthlog in Rice, Functions as an Ehd1 Repressor Through Interaction with Monocot-Specific CCT-Domain Protein Ghd7., Here, we report biological interactions between Ghd7 and Hd1, which together repress Early heading date 1 (Ehd1), a key floral inducer under non-inductive long-day (LD) conditions
- Ghd7, Hd1, Hd1,a CONSTANS Orthlog in Rice, Functions as an Ehd1 Repressor Through Interaction with Monocot-Specific CCT-Domain Protein Ghd7., These findings imply that Hd1, an evolutionally conserved transcriptional activator, can function as a strong transcriptional repressor within a monocot-specific flowering-time pathway through with Ghd7
- Ghd7, PHYA~OsPhyA, OsPhyA modulates rice flowering time mainly through OsGI under short days and Ghd7 under long days in the absence of phytochrome B., OsPhyA modulates rice flowering time mainly through OsGI under short days and Ghd7 under long days in the absence of phytochrome B.
- Ghd7, PHYA~OsPhyA, OsPhyA modulates rice flowering time mainly through OsGI under short days and Ghd7 under long days in the absence of phytochrome B., These results indicated that OsPhyA influences flowering time mainly by affecting the expression of OsGI under SD and Ghd7 under LD when phytochrome B is absent
- Ghd7, Se14, Se14, encoding a JmjC domain-containing protein, plays key roles in long-day suppression of rice flowering through the demethylation of H3K4me3 of RFT1., We found that Se14 is independent of the known photoperiod-sensitive genes, such as Hd1 and Ghd7, and is identical to Os03g0151300, which encodes a Jumonji C (JmjC) domain-containing protein
- Ghd7, ROC4, GL2-type homeobox gene Roc4 in rice promotes flowering time preferentially under long days by repressing Ghd7, GL2-type homeobox gene Roc4 in rice promotes flowering time preferentially under long days by repressing Ghd7
- Ghd7, ROC4, GL2-type homeobox gene Roc4 in rice promotes flowering time preferentially under long days by repressing Ghd7, Whereas constitutive overexpression of Roc4 in Dongjin japonica rice, which carries active Ghd7, also caused LD-preferential early flowering, its overexpression in Longjing27 rice, which is defective in functional Ghd7, did not produce the same result
- Ghd7, ROC4, GL2-type homeobox gene Roc4 in rice promotes flowering time preferentially under long days by repressing Ghd7, This confirmed that Roc4 regulates flowering time mainly through Ghd7
- Ghd7, ROC4, GL2-type homeobox gene Roc4 in rice promotes flowering time preferentially under long days by repressing Ghd7, All of these findings are evidence that Roc4 is an LD-preferential flowering enhancer that functions downstream of phytochromes and OsGI, but upstream of Ghd7
- Ghd2~OsK, Ghd7, Ghd2, a CONSTANS-like gene, confers drought sensitivity through regulation of senescence in rice., Here, we found that a CO-like gene, Ghd2 (Grain number, plant height, and heading date2), which can increase the yield potential under normal growth condition just like its homologue Ghd7, is involved in the regulation of leaf senescence and drought resistance
- Ghd7, OsCOL10, Flowering time regulation by the CONSTANS-Like gene OsCOL10., Moreover, we also showed that OsCOL10 acts downstream of Ghd7, a key LD-specific flowering repressor by reducing expression of Ehd1
- Ghd7, OsCOL10, Flowering time regulation by the CONSTANS-Like gene OsCOL10., Collectively, our finding identifies OsCOL10 functioning as a flowering-time repressor that links between Ghd7 and Ehd1 in rice
- Ghd7, OsCOL16, OsCOL16, encoding a CONSTANS-like protein, represses flowering by up-regulating Ghd7 expression in rice., OsCOL16, encoding a CONSTANS-like protein, represses flowering by up-regulating Ghd7 expression in rice.
- Ghd7, OsCOL16, OsCOL16, encoding a CONSTANS-like protein, represses flowering by up-regulating Ghd7 expression in rice., We determined that OsCOL16 up-regulates the expression of the floral repressor Ghd7, leading to down-regulation of the expression of Ehd1, Hd3a, and RFT1
- Ehd1, Ghd7, Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions., The interaction between proteins Ghd7 and Hd1 occurred through binding of the CCT domain of Ghd7 to the transcription-activating domain of Hd1, resulting in suppression of Ehd1 and florigen gene expression
- Ghd7, Hd1, Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions., Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions.
- Ghd7, Hd1, Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions., Comparative analysis of two sets of near isogenic lines of Hd1 in MH63 and ZS97 backgrounds indicated that the alternative functions of Hd1 in promoting or suppressing heading under LD are dependent on the previously cloned flowering repressor gene Ghd7
- Ghd7, Hd1, Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions., The interaction between proteins Ghd7 and Hd1 occurred through binding of the CCT domain of Ghd7 to the transcription-activating domain of Hd1, resulting in suppression of Ehd1 and florigen gene expression
- Ghd7, Hd1, Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions., These findings suggest that Hd1 alone essentially acts as a promoter of heading date, and the protein interaction between Ghd7 and Hd1 determines photoperiod sensitivity and integrated Hd1-mediated and Ehd1-mediated flowering pathways in rice