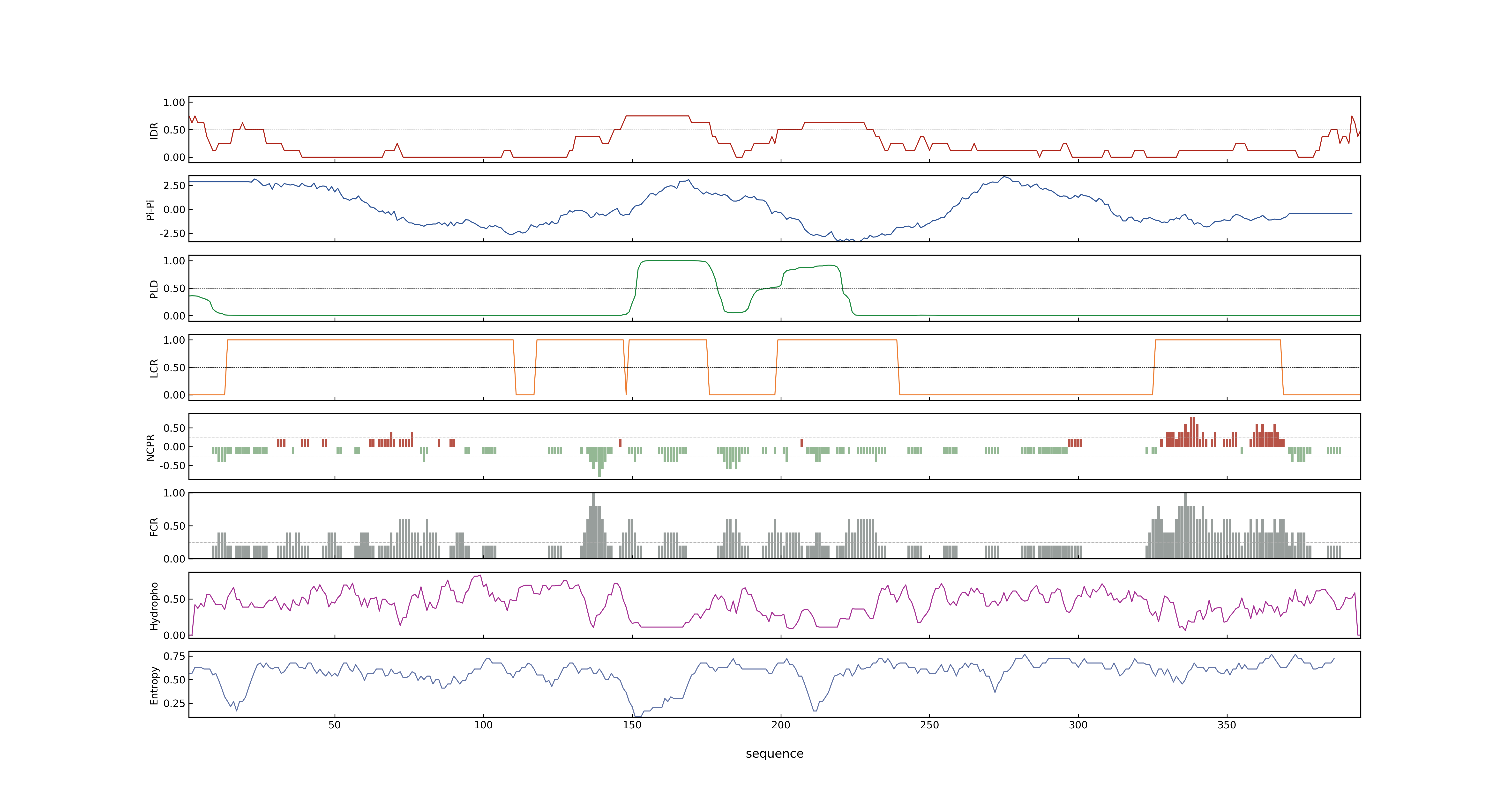
- Information
- Symbol: Hd1
- MSU: LOC_Os06g16370
- RAPdb: Os06g0275000
- PSP score
- LOC_Os06g16370.1: 0.5251
- PLAAC score
- LOC_Os06g16370.1: 20.721
- pLDDT score
- 63.1
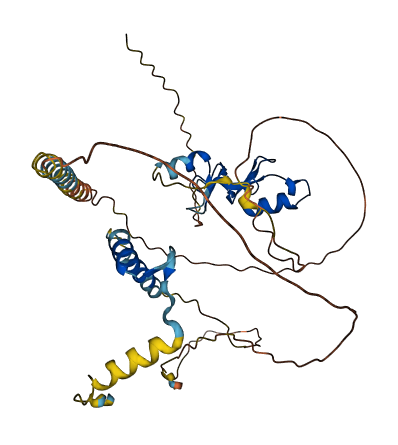
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os06g16370.1: 0.99983060
- MolPhase Result
- Publication
- Systematic identification of novel protein domain families associated with nuclear functions, 2002, Genome Res.
- An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, 2011, New Biotechnol.
- Genetic interactions involved in the inhibition of heading by heading date QTL, Hd2 in rice under long-day conditions, 2011, Theor Appl Genet.
- Identification of quantitative trait loci controlling heading date in rice using a high-density linkage map, 1997, TAG Theoretical and Applied Genetics.
- Fine mapping of quantitative trait loci Hd-1 , Hd-2 and Hd-3 , controlling heading date of rice, as single Mendelian factors, 1998, TAG Theoretical and Applied Genetics.
- Characterization and detection of epistatic interactions of 3 QTLs, Hd1 , Hd2 , and Hd3, controlling heading date in rice using nearly isogenic lines, 2000, TAG Theoretical and Applied Genetics.
- Heading date gene, dth3 controlled late flowering in O. Glaberrima Steud. by down-regulating Ehd1, 2011, Plant Cell Rep.
- OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice, 2008, Planta.
- Functional characterization of rice OsDof12, 2009, Planta.
- Phytochrome B regulates Heading date 1 Hd1-mediated expression of rice florigen Hd3a and critical day length in rice, 2011, Mol Genet Genomics.
- Epistasis among the three major flowering time genes in rice: coordinate changes of photoperiod sensitivity, basic vegetative growth and optimum photoperiod, 2007, Euphytica.
- Interactive effects of two heading-time loci, Se1 and Ef1, on pre-flowering developmental phases in rice Oryza sativa L., 2002, Euphytica.
- Heading date 1 Hd1, an ortholog of Arabidopsis CONSTANS, is a possible target of human selection during domestication to diversify flowering times of cultivated rice, 2011, Genes Genet Syst.
- A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice, 2009, Development.
- OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, 2009, Plant Cell Environ.
- Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 SOC1/AGL20 ortholog in rice, 2004, Plant J.
- Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering, 2011, Plant J.
- Footprints of natural and artificial selection for photoperiod pathway genes in Oryza, 2012, Plant J.
- The effect of the crosstalk between photoperiod and temperature on the heading-date in rice, 2009, PLoS One.
- Pleiotropism of the photoperiod-insensitive allele of Hd1 on heading date, plant height and yield traits in rice, 2012, PLoS One.
- Hd1, a Major Photoperiod Sensitivity Quantitative Trait Locus in Rice, Is Closely Related to the Arabidopsis Flowering Time Gene CONSTANS, 2000, The Plant Cell Online.
- Suppression of the floral activator Hd3a is the principal cause of the night break effect in rice, 2005, Plant Cell.
- Identification of dynamin as an interactor of rice GIGANTEA by tandem affinity purification TAP, 2008, Plant Cell Physiol.
- Flowering time genes Heading date 1 and Early heading date 1 together control panicle development in rice, 2011, Plant Cell Physiol.
- Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1, 2008, Plant Physiol.
- DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously, 2010, Plant Physiol.
- Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date3, 2014, Plant Physiol.
- Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice, 2009, Proc Natl Acad Sci U S A.
- Breeding strategies for optimum heading date using genotypic information in rice, 2009, Molecular Breeding.
- SPIN1, a K homology domain protein negatively regulated and ubiquitinated by the E3 ubiquitin ligase SPL11, is involved in flowering time control in rice, 2008, Plant Cell.
- Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response, 2013, Plant J.
- Transcriptional and Post-transcriptional Mechanisms Limit Heading Date 1 Hd1 Function to Adapt Rice to High Latitudes., 2017, PLoS Genet.
- Genbank accession number
- AB041837
- AB041838
- AB041839
- AB041840
- AB041841
- AB041842
- AP003044
- AB041837
- AB041838
- AB041839
- AB041840
- AB041841
- AB041842
- AP003044
- AB041837
- AB041838
- AB041839
- AB041840
- AB041841
- AB041842
- AP003044
- AB041837
- AB041838
- AB041839
- AB041840
- AB041841
- AB041842
- AP003044
- AB041837
- AB041838
- AB041839
- AB041840
- AB041841
- AB041842
- AP003044
- AB041837
- AB041838
- AB041839
- AB041840
- AB041841
- AB041842
- AP003044
- AB041837
- AB041838
- AB041839
- AB041840
- AB041841
- AB041842
- AP003044
- Key message
- Spin1 overexpression causes late flowering independently of daylength; expression analyses of flowering marker genes in these lines suggested that SPIN1 represses flowering by downregulating the flowering promoter gene Heading date3a (Hd3a) via Hd1-dependent mechanisms in short days and by targeting Hd1-independent factors in long days
- The CONSTANS (CO) and Heading date 1 (Hd1) genes are known to be central integrators of the photoperiod pathway in Arabidopsis and rice, respectively
- The expression of Hd3a and FTL decreased in these transgenic plants, whereas the expression of Hd1, Early heading date 1 (Ehd1), OsMADS51, and OsMADS50 did not significantly change
- RT-PCR analyses of the OsMADS50 KO and ubiquitin (ubi):OsMADS50 plants showed that OsMADS50 is an upstream regulator of OsMADS1, OsMADS14, OsMADS15, OsMADS18, and Hd (Heading date)3a, but works either parallel with or downstream of Hd1 and O
- Therefore, two key flowering time genes, Hd1 and Ehd1, can control panicle development in rice; this may affect crop yields in the field through florigen expression in leaf
- Meanwhile, the transcription of DTH8 has been proved to be independent of Ghd7 and Hd1, and the natural mutation of this gene caused weak photoperiod sensitivity and shorter plant height
- In addition, QTLs near Hd2, Hd16, and Ghd7, which are involved in inhibition of heading under long-day conditions, function in the same pathway that controls heading date
- The results revealed that the combination of Heading-date 1 (Hd1) and Early heading date 1 (Ehd1) can reduce the number of primary branches in a panicle, resulting in smaller spikelet numbers per panicle; this occurs independently of the control of flowering time
- Flowering genes downstream of OsPRR1 such as OsGI and Hd1 were down regulated in the A654 mutant
- Together, these results suggest that overexpression of OsLF might repress expression of OsGI and Hd1 by competing with OsPRR1 in interacting with OsPIL13 and OsPIL15 and thus induce late flowering
- A single NB strongly suppressed the mRNA of Hd3a, a homolog of Arabidopsis thaliana FLOWERING LOCUS T (FT), whereas the mRNAs of OsGI and Hd1 were not affected
- Since the photoperiod-insensitive allele of Hd1 confers a long vegetative phase, it is a good candidate for breeding rice varieties with high yielding potential for low latitudes
- The RNA levels of Heading date 3a (Hd3a), encoding a floral activator, are highly correlated with flowering time, and there is a high degree of polymorphism in the Heading date 1 (Hd1) protein, which is a major regulator of Hd3a expression
- Functional and nonfunctional alleles of Hd1 are associated with early and late flowering, respectively, suggesting that Hd1 is a major determinant of variation in flowering time of cultivated rice
- We also found that the type of Hd3a promoter and the level of Ehd1 expression contribute to the diversity in flowering time and Hd3a expression level
- Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice
- Map-based cloning revealed that the rice flowering-time quantitative trait locus (QTL) Heading date 16 (Hd16) encodes a casein kinase-I protein
- Heading date 1 (Hd1), a regulator of the florigen gene Hd3a, is one of the main factors used to generate diversity in flowering
- Loss-of-function alleles of Hd1 are common in cultivated rice and cause the diversity of flowering time
- In contrast with Hd3a, which has been highly conserved, Hd1 may have undergone human selection to diversify the flowering times of rice during domestication or the early stage of the cultivation period
- Heading date 1 (Hd1), an ortholog of Arabidopsis CONSTANS, is a possible target of human selection during domestication to diversify flowering times of cultivated rice
- We further revealed that Hd1 and/or Ehd1 caused up-regulation of Terminal Flower 1-like genes and precocious expression of panicle formation-related genes at shoot apical meristems during panicle development
- The comparison of the nucleotide sequences suggested that Ef1 is the same as Early heading date 1 (Ehd1)
- The quantitative real-time PCR assay revealed that DTH8 could down-regulate the transcriptions of Ehd1 (for Early heading date1) and Hd3a (for Heading date3a; a rice ortholog of FLOWERING LOCUS T) under long-day conditions
- The Early heading date 1 (Ehd1) which promotes the RFT1, was up-regulated by DTH3 in both LD and SD conditions
- Heading date gene, dth3 controlled late flowering in O. Glaberrima Steud. by down-regulating Ehd1
- By using near-isogenic lines with functional or deficient alleles of several rice flowering-time genes, we observed significant digenetic interactions between Hd16 and four other flowering-time genes (Ghd7, Hd1, DTH8 and Hd2)
- These results demonstrate that Hd16 acts as an inhibitor in the rice flowering pathway by enhancing the photoperiod response as a result of the phosphorylation of Ghd7
- Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response
- Characterization and detection of epistatic interactions of 3 QTLs, Hd1 , Hd2 , and Hd3, controlling heading date in rice using nearly isogenic lines
- We grew four rice lines having different flowering time genotypes (hd1 ehd1, hd1 Ehd1, Hd1 ehd1 and Hd1 Ehd1) under distinct photoperiod conditions
- In addition, expression of the Hd3a and Rice Flowering-locus T 1 (RFT1) florigen genes was up-regulated in leaves of the Hd1 Ehd1 line at the time of the floral transition
- They were used to analyze the effects of Hd1 on heading date, plant height and yield traits
- Pleiotropism of the photoperiod-insensitive allele of Hd1 on heading date, plant height and yield traits in rice
- We examined the footprints of natural and artificial selections for four major genes of the photoperiod pathway, namely PHYTOCHROME B (PhyB), HEADING DATE 1 (Hd1), HEADING DATE 3a (Hd3a), and EARLY HEADING DATE 1 (Ehd1), by investigation of the patterns of nucleotide polymorphisms in cultivated and wild rice
- In rice, OsGI, Hd1 and Hd3a were identified as orthologs of GI, CO and FT, respectively, and are also important regulators of flowering
- Hd1, a Major Photoperiod Sensitivity Quantitative Trait Locus in Rice, Is Closely Related to the Arabidopsis Flowering Time Gene CONSTANS
- Many other features of the photoperiod genes revealed domestication signatures, which included high linkage disequilibrium (LD) within genes, the occurrence of frequent and recurrent non-functional Hd1 mutants in cultivated rice, crossovers between subtropical and tropical alleles of Hd1, and significant LD between Hd1 and Hd3a in japonica and indica
- Under SD conditions, flowering is promoted through the activation of FT-like genes (rice florigens) by Heading date 1 (Hd1, a rice CONSTANS homolog) and Early heading date 1 (Ehd1, with no ortholog in the Arabidopsis genome)
- On the other hand, under long-day (LD) conditions, flowering is delayed by the repressive function of Hd1 on FT-like genes and by downregulation of Ehd1 by the flowering repressor Ghd7 - a unique pathway in rice
- We report here that an early heading date 3 (ehd3) mutant flowered later than wild-type plants, particularly under LD conditions, regardless of the Hd1-deficient background
- Furthermore, Ehd3 ghd7 plants flowered earlier and show higher Ehd1 transcript levels than ehd3 ghd7 plants, suggesting a Ghd7-independent role of Ehd3 in the upregulation of Ehd1
- In rice, a short-day plant (SDP), the CO ortholog Heading date 1 (Hd1) regulates FT ortholog Hd3a, but regulation of Hd3a by Hd1 differs from that in Arabidopsis
- Phytochrome B regulates Heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice
- The non-functional Hd1 alleles found in cultivated rice may be selected during domestication, because they were not found or very rare in wild ancestral rice
- We discovered an early heading date2 (ehd2) mutant that shows extremely late flowering under both short- and long-day conditions in line with a background deficient in Heading date1 (Hd1), a rice CONSTANS ortholog that belongs to the conserved pathway
- The present study was carried out in rice to examine to what extent these three developmental components are modified by the three flowering time genes, Se1 (= Hd1), Ef1 and e1 (= m-Ef1), which are known to contribute to flowering time in temperate and tropical regions of rice cultivation
- The expression patterns of Hd1 and Hd3a were also analyzed in different photoperiod and temperature conditions, revealing that Hd1 mRNA levels displayed similar expression patterns for different photoperiod and temperature treatments, with high expression levels at night and reduced levels in the daytime
- In addition, Hd1 displayed a slightly higher expression level under long-day and low temperature conditions
- Since the two genes Se1 (= Hd1) and Ef1 (= Ehd1) are known to up-regulate the rice homolog of Arabidopsis FT, it is suggested that the detected epistasis may respond to diverse environments by modulating the CO/FT system conserved in flowering plants
- We propose that both positive (OsMADS50 and Ehd1) and negative (Hd1, phyB and Ghd7) regulators of RFT1 form a gene network that regulates LD flowering in rice
- Among these regulators, Ehd1, a rice-specific floral inducer, integrates multiple pathways to regulate RFT1, leading to flowering under appropriate photoperiod conditions
- 3-kb region covering heading date gene Hd1 were developed from the indica rice cross Zhenshan97 (ZS97)/Milyang 46 (MY46)
- Ehd1 and Hd3a can also be down-regulated by the photoperiodic flowering genes Ghd7 and Hd1 (a rice ortholog of CONSTANS)
- To assign the position of Ehd2 within the flowering pathway of rice, we compared transcript levels of previously isolated flowering-time genes, such as Ehd1, a member of the unique pathway, Hd3a, and Rice FT-like1 (RFT1; rice florigens), between the wild-type plants and the ehd2 mutants
- Severely reduced expression of these genes in ehd2 under both short- and long-day conditions suggests that Ehd2 acts as a flowering promoter mainly by up-regulating Ehd1 and by up-regulating the downstream Hd3a and RFT1 genes in the unique genetic network of photoperiodic flowering in rice
- Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1
- First, overexpression of Hd1 causes a delay in flowering under SD conditions and this effect requires phyB, suggesting that light modulates Hd1 control of Hd3a transcription
- We demonstrate that a histone fold domain scaffold formed by GRAIN YIELD, PLANT HEIGHT AND HEADING DATE 8 (Ghd8) and several NF-YC subunits can accommodate distinct proteins, including Hd1 and PSEUDO RESPONSE REGULATOR 37 (PRR37), and that the resulting OsNF-Y complex containing Hd1 can bind a specific sequence in the promoter of HEADING DATE 3A (Hd3a)
- Connection
- Hd1, OsLF, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Together, these results suggest that overexpression of OsLF might repress expression of OsGI and Hd1 by competing with OsPRR1 in interacting with OsPIL13 and OsPIL15 and thus induce late flowering
- Hd1, OsGI, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Flowering genes downstream of OsPRR1 such as OsGI and Hd1 were down regulated in the A654 mutant
- Hd1, OsGI, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Together, these results suggest that overexpression of OsLF might repress expression of OsGI and Hd1 by competing with OsPRR1 in interacting with OsPIL13 and OsPIL15 and thus induce late flowering
- Hd1, OsPRR1, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Flowering genes downstream of OsPRR1 such as OsGI and Hd1 were down regulated in the A654 mutant
- Hd1, OsPRR1, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Together, these results suggest that overexpression of OsLF might repress expression of OsGI and Hd1 by competing with OsPRR1 in interacting with OsPIL13 and OsPIL15 and thus induce late flowering
- Hd1, OsPIL13~OsPIL1, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Together, these results suggest that overexpression of OsLF might repress expression of OsGI and Hd1 by competing with OsPRR1 in interacting with OsPIL13 and OsPIL15 and thus induce late flowering
- Hd1, OsPIL15, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Together, these results suggest that overexpression of OsLF might repress expression of OsGI and Hd1 by competing with OsPRR1 in interacting with OsPIL13 and OsPIL15 and thus induce late flowering
- Ghd7, Hd1, Genetic interactions involved in the inhibition of heading by heading date QTL, Hd2 in rice under long-day conditions, Combinations of several QTLs near Hd1, Hd2, Ghd7, Hd5, and Hd16 were detected under these four conditions
- Hd1, Hd5~DTH8~Ghd8~OsHAP3H~LHD1~EF8~CAR8~OsNF-YB11, Genetic interactions involved in the inhibition of heading by heading date QTL, Hd2 in rice under long-day conditions, Combinations of several QTLs near Hd1, Hd2, Ghd7, Hd5, and Hd16 were detected under these four conditions
- CKI~EL1~Hd16, Hd1, Genetic interactions involved in the inhibition of heading by heading date QTL, Hd2 in rice under long-day conditions, Combinations of several QTLs near Hd1, Hd2, Ghd7, Hd5, and Hd16 were detected under these four conditions
- Hd1, OsCO3, OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice, The circadian expression pattern of OsCO3 mRNA oscillated in a different phase from Hd1 and was similar to that of OsCO3 pre-mRNA, suggesting that the diurnal expression pattern of OsCO3 transcripts may be regulated at the transcriptional level
- Ehd1, Hd1, OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice, The expression of Hd3a and FTL decreased in these transgenic plants, whereas the expression of Hd1, Early heading date 1 (Ehd1), OsMADS51, and OsMADS50 did not significantly change
- Hd1, Hd3a, OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice, The expression of Hd3a and FTL decreased in these transgenic plants, whereas the expression of Hd1, Early heading date 1 (Ehd1), OsMADS51, and OsMADS50 did not significantly change
- Hd1, OsMADS51~OsMADS65, OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice, The expression of Hd3a and FTL decreased in these transgenic plants, whereas the expression of Hd1, Early heading date 1 (Ehd1), OsMADS51, and OsMADS50 did not significantly change
- Hd1, OsMADS50~OsSOC1~DTH3, OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice, The expression of Hd3a and FTL decreased in these transgenic plants, whereas the expression of Hd1, Early heading date 1 (Ehd1), OsMADS51, and OsMADS50 did not significantly change
- Ehd1, Hd1, Functional characterization of rice OsDof12, In transgenic lines overexpressing OsDof12, the transcription levels of Hd3a and OsMADS14 were up-regulated under LD conditions but not SD conditions, whereas the expression of Hd1, OsMADS51, Ehd1 and OsGI did not change under LD and SD conditions
- Hd1, OsMADS51~OsMADS65, Functional characterization of rice OsDof12, In transgenic lines overexpressing OsDof12, the transcription levels of Hd3a and OsMADS14 were up-regulated under LD conditions but not SD conditions, whereas the expression of Hd1, OsMADS51, Ehd1 and OsGI did not change under LD and SD conditions
- Hd1, OsGI, Functional characterization of rice OsDof12, In transgenic lines overexpressing OsDof12, the transcription levels of Hd3a and OsMADS14 were up-regulated under LD conditions but not SD conditions, whereas the expression of Hd1, OsMADS51, Ehd1 and OsGI did not change under LD and SD conditions
- Hd1, Hd3a, Functional characterization of rice OsDof12, In transgenic lines overexpressing OsDof12, the transcription levels of Hd3a and OsMADS14 were up-regulated under LD conditions but not SD conditions, whereas the expression of Hd1, OsMADS51, Ehd1 and OsGI did not change under LD and SD conditions
- Hd1, OsMADS14, Functional characterization of rice OsDof12, In transgenic lines overexpressing OsDof12, the transcription levels of Hd3a and OsMADS14 were up-regulated under LD conditions but not SD conditions, whereas the expression of Hd1, OsMADS51, Ehd1 and OsGI did not change under LD and SD conditions
- Hd1, OsDof12~OsCDF1, Functional characterization of rice OsDof12, In transgenic lines overexpressing OsDof12, the transcription levels of Hd3a and OsMADS14 were up-regulated under LD conditions but not SD conditions, whereas the expression of Hd1, OsMADS51, Ehd1 and OsGI did not change under LD and SD conditions
- Hd1, PHYB~OsphyB, Phytochrome B regulates Heading date 1 Hd1-mediated expression of rice florigen Hd3a and critical day length in rice, First, overexpression of Hd1 causes a delay in flowering under SD conditions and this effect requires phyB, suggesting that light modulates Hd1 control of Hd3a transcription
- Hd1, Hd3a, Phytochrome B regulates Heading date 1 Hd1-mediated expression of rice florigen Hd3a and critical day length in rice, In rice, a short-day plant (SDP), the CO ortholog Heading date 1 (Hd1) regulates FT ortholog Hd3a, but regulation of Hd3a by Hd1 differs from that in Arabidopsis
- Hd1, Hd3a, Phytochrome B regulates Heading date 1 Hd1-mediated expression of rice florigen Hd3a and critical day length in rice, First, overexpression of Hd1 causes a delay in flowering under SD conditions and this effect requires phyB, suggesting that light modulates Hd1 control of Hd3a transcription
- Hd1, Hd3a, Phytochrome B regulates Heading date 1 Hd1-mediated expression of rice florigen Hd3a and critical day length in rice, Phytochrome B regulates Heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice
- Ehd1, Hd1, Epistasis among the three major flowering time genes in rice: coordinate changes of photoperiod sensitivity, basic vegetative growth and optimum photoperiod, Since the two genes Se1 (= Hd1) and Ef1 (= Ehd1) are known to up-regulate the rice homolog of Arabidopsis FT, it is suggested that the detected epistasis may respond to diverse environments by modulating the CO/FT system conserved in flowering plants
- Ehd1, Hd1, A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice, We propose that both positive (OsMADS50 and Ehd1) and negative (Hd1, phyB and Ghd7) regulators of RFT1 form a gene network that regulates LD flowering in rice
- Hd1, OsMADS50~OsSOC1~DTH3, A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice, We propose that both positive (OsMADS50 and Ehd1) and negative (Hd1, phyB and Ghd7) regulators of RFT1 form a gene network that regulates LD flowering in rice
- Ghd7, Hd1, A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice, We propose that both positive (OsMADS50 and Ehd1) and negative (Hd1, phyB and Ghd7) regulators of RFT1 form a gene network that regulates LD flowering in rice
- Hd1, PHYB~OsphyB, A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice, We propose that both positive (OsMADS50 and Ehd1) and negative (Hd1, phyB and Ghd7) regulators of RFT1 form a gene network that regulates LD flowering in rice
- Hd1, RFT1, A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice, We propose that both positive (OsMADS50 and Ehd1) and negative (Hd1, phyB and Ghd7) regulators of RFT1 form a gene network that regulates LD flowering in rice
- Hd1, Hd6~CK2, OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, On the other hand, mRNA levels of OsGI, Hd1, OsId1, OsDof12, Ghd7, Hd6 and SE5 were unchanged
- Hd1, OsDof12~OsCDF1, OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, On the other hand, mRNA levels of OsGI, Hd1, OsId1, OsDof12, Ghd7, Hd6 and SE5 were unchanged
- Hd1, OsGI, OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, On the other hand, mRNA levels of OsGI, Hd1, OsId1, OsDof12, Ghd7, Hd6 and SE5 were unchanged
- Ghd7, Hd1, OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, On the other hand, mRNA levels of OsGI, Hd1, OsId1, OsDof12, Ghd7, Hd6 and SE5 were unchanged
- Hd1, OsMADS1~LHS1~AFO, Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 SOC1/AGL20 ortholog in rice, RT-PCR analyses of the OsMADS50 KO and ubiquitin (ubi):OsMADS50 plants showed that OsMADS50 is an upstream regulator of OsMADS1, OsMADS14, OsMADS15, OsMADS18, and Hd (Heading date)3a, but works either parallel with or downstream of Hd1 and O
- Hd1, OsMADS14, Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 SOC1/AGL20 ortholog in rice, RT-PCR analyses of the OsMADS50 KO and ubiquitin (ubi):OsMADS50 plants showed that OsMADS50 is an upstream regulator of OsMADS1, OsMADS14, OsMADS15, OsMADS18, and Hd (Heading date)3a, but works either parallel with or downstream of Hd1 and O
- Hd1, OsMADS15~DEP, Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 SOC1/AGL20 ortholog in rice, RT-PCR analyses of the OsMADS50 KO and ubiquitin (ubi):OsMADS50 plants showed that OsMADS50 is an upstream regulator of OsMADS1, OsMADS14, OsMADS15, OsMADS18, and Hd (Heading date)3a, but works either parallel with or downstream of Hd1 and O
- Hd1, OsMADS50~OsSOC1~DTH3, Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 SOC1/AGL20 ortholog in rice, RT-PCR analyses of the OsMADS50 KO and ubiquitin (ubi):OsMADS50 plants showed that OsMADS50 is an upstream regulator of OsMADS1, OsMADS14, OsMADS15, OsMADS18, and Hd (Heading date)3a, but works either parallel with or downstream of Hd1 and O
- Hd1, OsMADS18, Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 SOC1/AGL20 ortholog in rice, RT-PCR analyses of the OsMADS50 KO and ubiquitin (ubi):OsMADS50 plants showed that OsMADS50 is an upstream regulator of OsMADS1, OsMADS14, OsMADS15, OsMADS18, and Hd (Heading date)3a, but works either parallel with or downstream of Hd1 and O
- Ghd7, Hd1, Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering, On the other hand, under long-day (LD) conditions, flowering is delayed by the repressive function of Hd1 on FT-like genes and by downregulation of Ehd1 by the flowering repressor Ghd7 - a unique pathway in rice
- Ehd1, Hd1, Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering, On the other hand, under long-day (LD) conditions, flowering is delayed by the repressive function of Hd1 on FT-like genes and by downregulation of Ehd1 by the flowering repressor Ghd7 - a unique pathway in rice
- Hd1, PHYB~OsphyB, Footprints of natural and artificial selection for photoperiod pathway genes in Oryza, We examined the footprints of natural and artificial selections for four major genes of the photoperiod pathway, namely PHYTOCHROME B (PhyB), HEADING DATE 1 (Hd1), HEADING DATE 3a (Hd3a), and EARLY HEADING DATE 1 (Ehd1), by investigation of the patterns of nucleotide polymorphisms in cultivated and wild rice
- Hd1, Hd3a, Footprints of natural and artificial selection for photoperiod pathway genes in Oryza, We examined the footprints of natural and artificial selections for four major genes of the photoperiod pathway, namely PHYTOCHROME B (PhyB), HEADING DATE 1 (Hd1), HEADING DATE 3a (Hd3a), and EARLY HEADING DATE 1 (Ehd1), by investigation of the patterns of nucleotide polymorphisms in cultivated and wild rice
- Hd1, Hd3a, Footprints of natural and artificial selection for photoperiod pathway genes in Oryza, Many other features of the photoperiod genes revealed domestication signatures, which included high linkage disequilibrium (LD) within genes, the occurrence of frequent and recurrent non-functional Hd1 mutants in cultivated rice, crossovers between subtropical and tropical alleles of Hd1, and significant LD between Hd1 and Hd3a in japonica and indica
- Hd1, Hd3a, The effect of the crosstalk between photoperiod and temperature on the heading-date in rice, The expression patterns of Hd1 and Hd3a were also analyzed in different photoperiod and temperature conditions, revealing that Hd1 mRNA levels displayed similar expression patterns for different photoperiod and temperature treatments, with high expression levels at night and reduced levels in the daytime
- Hd1, Hd3a, Suppression of the floral activator Hd3a is the principal cause of the night break effect in rice, A single NB strongly suppressed the mRNA of Hd3a, a homolog of Arabidopsis thaliana FLOWERING LOCUS T (FT), whereas the mRNAs of OsGI and Hd1 were not affected
- Hd1, Hd3a, Identification of dynamin as an interactor of rice GIGANTEA by tandem affinity purification TAP, In rice, OsGI, Hd1 and Hd3a were identified as orthologs of GI, CO and FT, respectively, and are also important regulators of flowering
- Hd1, OsGI, Identification of dynamin as an interactor of rice GIGANTEA by tandem affinity purification TAP, In rice, OsGI, Hd1 and Hd3a were identified as orthologs of GI, CO and FT, respectively, and are also important regulators of flowering
- Ehd1, Hd1, Flowering time genes Heading date 1 and Early heading date 1 together control panicle development in rice, We grew four rice lines having different flowering time genotypes (hd1 ehd1, hd1 Ehd1, Hd1 ehd1 and Hd1 Ehd1) under distinct photoperiod conditions
- Ehd1, Hd1, Flowering time genes Heading date 1 and Early heading date 1 together control panicle development in rice, In addition, expression of the Hd3a and Rice Flowering-locus T 1 (RFT1) florigen genes was up-regulated in leaves of the Hd1 Ehd1 line at the time of the floral transition
- Ehd1, Hd1, Flowering time genes Heading date 1 and Early heading date 1 together control panicle development in rice, We further revealed that Hd1 and/or Ehd1 caused up-regulation of Terminal Flower 1-like genes and precocious expression of panicle formation-related genes at shoot apical meristems during panicle development
- Ehd1, Hd1, Flowering time genes Heading date 1 and Early heading date 1 together control panicle development in rice, Therefore, two key flowering time genes, Hd1 and Ehd1, can control panicle development in rice; this may affect crop yields in the field through florigen expression in leaf
- Hd1, Hd3a, Flowering time genes Heading date 1 and Early heading date 1 together control panicle development in rice, In addition, expression of the Hd3a and Rice Flowering-locus T 1 (RFT1) florigen genes was up-regulated in leaves of the Hd1 Ehd1 line at the time of the floral transition
- Hd1, RFT1, Flowering time genes Heading date 1 and Early heading date 1 together control panicle development in rice, In addition, expression of the Hd3a and Rice Flowering-locus T 1 (RFT1) florigen genes was up-regulated in leaves of the Hd1 Ehd1 line at the time of the floral transition
- Ehd2~RID1, Hd1, Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1, We discovered an early heading date2 (ehd2) mutant that shows extremely late flowering under both short- and long-day conditions in line with a background deficient in Heading date1 (Hd1), a rice CONSTANS ortholog that belongs to the conserved pathway
- Hd1, Hd5~DTH8~Ghd8~OsHAP3H~LHD1~EF8~CAR8~OsNF-YB11, DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously, Meanwhile, the transcription of DTH8 has been proved to be independent of Ghd7 and Hd1, and the natural mutation of this gene caused weak photoperiod sensitivity and shorter plant height
- Ehd1, Hd1, DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously, Ehd1 and Hd3a can also be down-regulated by the photoperiodic flowering genes Ghd7 and Hd1 (a rice ortholog of CONSTANS)
- Ghd7, Hd1, DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously, Ehd1 and Hd3a can also be down-regulated by the photoperiodic flowering genes Ghd7 and Hd1 (a rice ortholog of CONSTANS)
- Ghd7, Hd1, DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously, Meanwhile, the transcription of DTH8 has been proved to be independent of Ghd7 and Hd1, and the natural mutation of this gene caused weak photoperiod sensitivity and shorter plant height
- Hd1, Hd3a, DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously, Ehd1 and Hd3a can also be down-regulated by the photoperiodic flowering genes Ghd7 and Hd1 (a rice ortholog of CONSTANS)
- Ehd1, Hd1, Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice, We also found that the type of Hd3a promoter and the level of Ehd1 expression contribute to the diversity in flowering time and Hd3a expression level
- Ehd1, Hd1, Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice, Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice
- Hd1, Hd3a, Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice, The RNA levels of Heading date 3a (Hd3a), encoding a floral activator, are highly correlated with flowering time, and there is a high degree of polymorphism in the Heading date 1 (Hd1) protein, which is a major regulator of Hd3a expression
- Hd1, Hd3a, Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice, We also found that the type of Hd3a promoter and the level of Ehd1 expression contribute to the diversity in flowering time and Hd3a expression level
- Hd1, Hd3a, Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice, Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice
- Hd1, Hd3a, SPIN1, a K homology domain protein negatively regulated and ubiquitinated by the E3 ubiquitin ligase SPL11, is involved in flowering time control in rice, Spin1 overexpression causes late flowering independently of daylength; expression analyses of flowering marker genes in these lines suggested that SPIN1 represses flowering by downregulating the flowering promoter gene Heading date3a (Hd3a) via Hd1-dependent mechanisms in short days and by targeting Hd1-independent factors in long days
- Hd1, SPIN1, SPIN1, a K homology domain protein negatively regulated and ubiquitinated by the E3 ubiquitin ligase SPL11, is involved in flowering time control in rice, Spin1 overexpression causes late flowering independently of daylength; expression analyses of flowering marker genes in these lines suggested that SPIN1 represses flowering by downregulating the flowering promoter gene Heading date3a (Hd3a) via Hd1-dependent mechanisms in short days and by targeting Hd1-independent factors in long days
- Hd1, Hd5~DTH8~Ghd8~OsHAP3H~LHD1~EF8~CAR8~OsNF-YB11, Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response, By using near-isogenic lines with functional or deficient alleles of several rice flowering-time genes, we observed significant digenetic interactions between Hd16 and four other flowering-time genes (Ghd7, Hd1, DTH8 and Hd2)
- Ghd7, Hd1, Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response, By using near-isogenic lines with functional or deficient alleles of several rice flowering-time genes, we observed significant digenetic interactions between Hd16 and four other flowering-time genes (Ghd7, Hd1, DTH8 and Hd2)
- CKI~EL1~Hd16, Hd1, Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response, By using near-isogenic lines with functional or deficient alleles of several rice flowering-time genes, we observed significant digenetic interactions between Hd16 and four other flowering-time genes (Ghd7, Hd1, DTH8 and Hd2)
- Ghd7, Hd1, Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice., Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice.
- Ghd7, Hd1, Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice., Map-based cloning demonstrated that the gene largely affecting the interaction between Ghd7 and Ghd8 was Hd1
- Ghd7, Hd1, Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice., Natural variation analysis showed that a combination of loss-of-function alleles of Ghd7, Ghd8 and Hd1 contributes to the expansion of rice cultivars to higher latitudes; by contrast, a combination of pre-existing strong alleles of Ghd7, Ghd8 and functional Hd1 (referred as SSF) is exclusively found where ancestral Asian cultivars originated
- Ghd7, Hd1, Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice., Our results indicate that the combinations of Ghd7, Ghd8 and Hd1 largely define the ecogeographical adaptation and yield potential in rice cultivars
- Hd1, OsHIP1~HAF1, The RING-Finger Ubiquitin Ligase HAF1 Mediates Heading date 1 Degradation during Photoperiodic Flowering in Rice., Here, we identify a C3HC4 RING domain-containing E3 ubiquitin ligase, Heading date Associated Factor 1 (HAF1), which physically interacts with Hd1
- Hd1, OsHIP1~HAF1, The RING-Finger Ubiquitin Ligase HAF1 Mediates Heading date 1 Degradation during Photoperiodic Flowering in Rice., HAF1 mediates ubiquitination and targets Hd1 for degradation via the 26S proteasome-dependent pathway
- Hd1, OsHIP1~HAF1, The RING-Finger Ubiquitin Ligase HAF1 Mediates Heading date 1 Degradation during Photoperiodic Flowering in Rice., In addition, the haf1 hd1 double mutant headed as late as hd1 plants under short-day conditions but exhibited a heading date similar to haf1 under long-day conditions, thus indicating that HAF1 may determine heading date mainly through Hd1 under short-day conditions
- Hd1, OsHIP1~HAF1, The RING-Finger Ubiquitin Ligase HAF1 Mediates Heading date 1 Degradation during Photoperiodic Flowering in Rice., Moreover, high levels of Hd1 accumulate in haf1
- Hd1, OsHIP1~HAF1, The RING-Finger Ubiquitin Ligase HAF1 Mediates Heading date 1 Degradation during Photoperiodic Flowering in Rice., Our results suggest that HAF1 is essential to precise modulation of the timing of Hd1 accumulation during the photoperiod response in rice
- Hd1, OsHAL3, OsHAL3, a new component interacts with the floral regulator Hd1 to activate flowering in rice., OsHAL3, a new component interacts with the floral regulator Hd1 to activate flowering in rice.
- Hd1, OsHAL3, OsHAL3, a new component interacts with the floral regulator Hd1 to activate flowering in rice., OsHAL3 and Hd1 colocalized to the nucleus in vivo and interacted physically in the dark, but the interaction was inhibited by white or blue light
- Hd1, OsHAL3, OsHAL3, a new component interacts with the floral regulator Hd1 to activate flowering in rice., We conclude that OsHAL3, a novel light responsive protein, plays an essential role in photoperiodic control of flowering time in rice, which is probably mediated by forming a complex with Hd1
- Ehd1, Hd1, Both Hd1 and Ehd1 are important for artificial selection of flowering time in cultivated rice., Both Hd1 and Ehd1 are important for artificial selection of flowering time in cultivated rice.
- Ehd1, Hd1, Both Hd1 and Ehd1 are important for artificial selection of flowering time in cultivated rice., These varieties contain loss-of-function of two important flowering-time related genes, Heading date 1 (Hd1) and Early heading date 1 (Ehd1), and are mainly from a mega variety, Taichung 65
- Ehd1, Hd1, Both Hd1 and Ehd1 are important for artificial selection of flowering time in cultivated rice., Both Hd1 and Ehd1 may be important during artificial selection for flowering time, especially in a subtropical region such as Taiwan
- Hd1, HDR1, The Oryza sativa Regulator HDR1 Associates with the Kinase OsK4 to Control Photoperiodic Flowering., We determined that HDR1 is a novel suppressor of flowering that upregulates Hd1 and downregulates Ehd1, leading to the downregulation of Hd3a and RFT1 under LDs
- Hd1, HDR1, The Oryza sativa Regulator HDR1 Associates with the Kinase OsK4 to Control Photoperiodic Flowering., OsK4 acts similarly to HDR1, suppressing flowering by upregulating Hd1 and downregulating Ehd1 under LDs, and OsK4 can phosphorylate HD1 with HDR1 presents
- Hd1, SnRK1B~OsK4, The Oryza sativa Regulator HDR1 Associates with the Kinase OsK4 to Control Photoperiodic Flowering., OsK4 acts similarly to HDR1, suppressing flowering by upregulating Hd1 and downregulating Ehd1 under LDs, and OsK4 can phosphorylate HD1 with HDR1 presents
- Ghd7, Hd1, Hd1,a CONSTANS Orthlog in Rice, Functions as an Ehd1 Repressor Through Interaction with Monocot-Specific CCT-Domain Protein Ghd7., Here, we report biological interactions between Ghd7 and Hd1, which together repress Early heading date 1 (Ehd1), a key floral inducer under non-inductive long-day (LD) conditions
- Ghd7, Hd1, Hd1,a CONSTANS Orthlog in Rice, Functions as an Ehd1 Repressor Through Interaction with Monocot-Specific CCT-Domain Protein Ghd7., These findings imply that Hd1, an evolutionally conserved transcriptional activator, can function as a strong transcriptional repressor within a monocot-specific flowering-time pathway through with Ghd7
- Hd1, OsBBX14, OsBBX14 delays heading date by repressing florigen gene expression under long and short-day conditions in rice, Thus, OsBBX14 acts as a floral repressor by promoting Hd1 expression under LD conditions, probably because of crosstalk with the circadian clock
- Hd1, OsBBX14, OsBBX14 delays heading date by repressing florigen gene expression under long and short-day conditions in rice, Under SD conditions, Ehd1 expression was reduced in OsBBX14-OX lines, but Hd1 and circadian clock gene expressions were unaffected, indicating that OsBBX14 acts as a repressor of Ehd1
- Hd1, Se14, Se14, encoding a JmjC domain-containing protein, plays key roles in long-day suppression of rice flowering through the demethylation of H3K4me3 of RFT1., We found that Se14 is independent of the known photoperiod-sensitive genes, such as Hd1 and Ghd7, and is identical to Os03g0151300, which encodes a Jumonji C (JmjC) domain-containing protein
- Hd1, OsHAPL1, The OsHAPL1-DTH8-Hd1 complex functions as the transcription regulator to repress heading date in rice., OsHAPL1 can physically interact with Days To Heading on chromosome 8 (DTH8), which physically interacts with Heading date 1 (Hd1) both in vitro and in vivo OsHAPL1 forms a complex with DTH8 and Hd1 in Escherichia coli OsHAPL1, DTH8, and Hd1 physically interact with the HAP complex, and also with general transcription factors in yeast (Saccharomyces cerevisiae)
- Hd1, OsHAPL1, The OsHAPL1-DTH8-Hd1 complex functions as the transcription regulator to repress heading date in rice., We propose that OsHAPL1 functions as a transcriptional regulator and, together with DTH8, Hd1, the HAP complex, and general transcription factors, regulates the expression of target genes and then affects heading date by influencing the expression of Hd3a and RFT1 through Ehd1
- Hd1, Hd3a, Transcriptional and Post-transcriptional Mechanisms Limit Heading Date 1 Hd1 Function to Adapt Rice to High Latitudes., We demonstrate that a histone fold domain scaffold formed by GRAIN YIELD, PLANT HEIGHT AND HEADING DATE 8 (Ghd8) and several NF-YC subunits can accommodate distinct proteins, including Hd1 and PSEUDO RESPONSE REGULATOR 37 (PRR37), and that the resulting OsNF-Y complex containing Hd1 can bind a specific sequence in the promoter of HEADING DATE 3A (Hd3a)
- Hd1, Hd5~DTH8~Ghd8~OsHAP3H~LHD1~EF8~CAR8~OsNF-YB11, Transcriptional and Post-transcriptional Mechanisms Limit Heading Date 1 Hd1 Function to Adapt Rice to High Latitudes., We demonstrate that a histone fold domain scaffold formed by GRAIN YIELD, PLANT HEIGHT AND HEADING DATE 8 (Ghd8) and several NF-YC subunits can accommodate distinct proteins, including Hd1 and PSEUDO RESPONSE REGULATOR 37 (PRR37), and that the resulting OsNF-Y complex containing Hd1 can bind a specific sequence in the promoter of HEADING DATE 3A (Hd3a)
- Hd1, OsMADS7~OsMADS45, Ectopic expression of OsMADS45 activates the upstream genes Hd3a and RFT1 at an early development stage causing early flowering in rice., We introduce an OsMADS45 overexpression construct Ubi:OsMADS45 into TNG67 plants (an Hd1 (Heading date 1) and Ehd1 (Early heading date 1) defective rice cultivar grown in Taiwan), and we analyzed the expression patterns of various floral regulators to understand the regulation pathways affected by OsMADS45 expression
- Hd1, OsMADS7~OsMADS45, Ectopic expression of OsMADS45 activates the upstream genes Hd3a and RFT1 at an early development stage causing early flowering in rice., OsMADS45 overexpression did not alter the oscillating rhythm of the examined floral regulatory genes but advanced (by approximately 20<U+00A0>days) the up-regulate of two florigens, Hd3a (Heading Date 3a) and RFT1 (RICE FLOWERING LOCUS T1) and suppressed the expression of Hd1 at the juvenile stage
- Hd1, OsMADS7~OsMADS45, Ectopic expression of OsMADS45 activates the upstream genes Hd3a and RFT1 at an early development stage causing early flowering in rice., OsMADS45 overexpression did not influence other floral regulator genes upstream of Hd1 and Ehd1, such as OsGI (OsGIGANTEA), Ehd2/Osld1/RID1 and OsMADS50
- Hd1, Hd5~DTH8~Ghd8~OsHAP3H~LHD1~EF8~CAR8~OsNF-YB11, The DTH8-Hd1 module mediates day length-dependent regulation of rice flowering., Here, we demonstrate that the repression of flowering in LD mediated by Hd1 is dependent on the transcription factor DAYS TO HEADING 8 (DTH8)
- Hd1, Hd5~DTH8~Ghd8~OsHAP3H~LHD1~EF8~CAR8~OsNF-YB11, The DTH8-Hd1 module mediates day length-dependent regulation of rice flowering., Loss of DTH8 function results in the activation of Hd3a by Hd1, leading to early flowering
- Hd1, Hd5~DTH8~Ghd8~OsHAP3H~LHD1~EF8~CAR8~OsNF-YB11, The DTH8-Hd1 module mediates day length-dependent regulation of rice flowering., We also show that Hd1 directly interacts with DTH8 and that the formation of the DTH8-Hd1 complex is necessary for the transcriptional repression of Hd3a by Hd1 in LD
- Hd1, Hd5~DTH8~Ghd8~OsHAP3H~LHD1~EF8~CAR8~OsNF-YB11, The DTH8-Hd1 module mediates day length-dependent regulation of rice flowering., We thus detected a switch in function of Hd1 mediated by DTH8 in LD rather than in SD
- Hd1, Hd5~DTH8~Ghd8~OsHAP3H~LHD1~EF8~CAR8~OsNF-YB11, The DTH8-Hd1 module mediates day length-dependent regulation of rice flowering., H3K27 trimethylation increases at Hd3a in the presence the DTH8-Hd1 complex, while Hd1 attenuates the H3K27me3 level in Hd3a when DTH8 function is lost
- Hd1, Hd5~DTH8~Ghd8~OsHAP3H~LHD1~EF8~CAR8~OsNF-YB11, The DTH8-Hd1 module mediates day length-dependent regulation of rice flowering., Therefore, our findings establish that, in response to day length, DTH8 plays a critical role in mediating Hd1 regulation of Hd3a transcription through the DTH8-Hd1 module to shape epigenetic marks in photoperiodic flowering
- Ghd7, Hd1, Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions., Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions.
- Ghd7, Hd1, Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions., Comparative analysis of two sets of near isogenic lines of Hd1 in MH63 and ZS97 backgrounds indicated that the alternative functions of Hd1 in promoting or suppressing heading under LD are dependent on the previously cloned flowering repressor gene Ghd7
- Ghd7, Hd1, Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions., The interaction between proteins Ghd7 and Hd1 occurred through binding of the CCT domain of Ghd7 to the transcription-activating domain of Hd1, resulting in suppression of Ehd1 and florigen gene expression
- Ghd7, Hd1, Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions., These findings suggest that Hd1 alone essentially acts as a promoter of heading date, and the protein interaction between Ghd7 and Hd1 determines photoperiod sensitivity and integrated Hd1-mediated and Ehd1-mediated flowering pathways in rice
Prev Next