- Information
- Symbol: IDEF2
- MSU: LOC_Os05g35170
- RAPdb: Os05g0426200
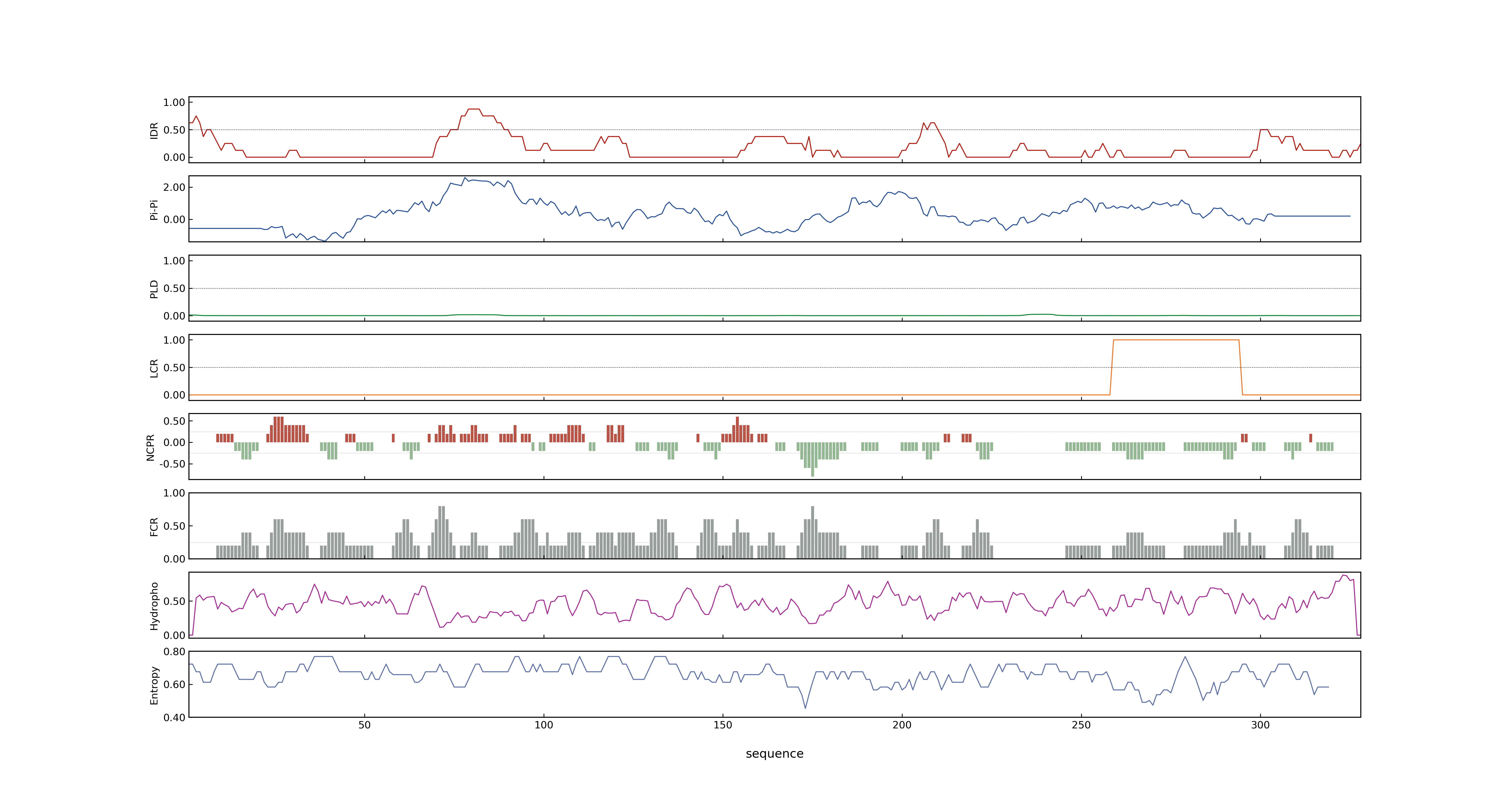
- PSP score
- LOC_Os05g35170.4: 0.0116
- LOC_Os05g35170.2: 0.0928
- LOC_Os05g35170.1: 0.0928
- LOC_Os05g35170.5: 0.0254
- LOC_Os05g35170.3: 0.0543
- PLAAC score
- LOC_Os05g35170.4: 0
- LOC_Os05g35170.2: 0
- LOC_Os05g35170.1: 0
- LOC_Os05g35170.5: 0
- LOC_Os05g35170.3: 0
- pLDDT score
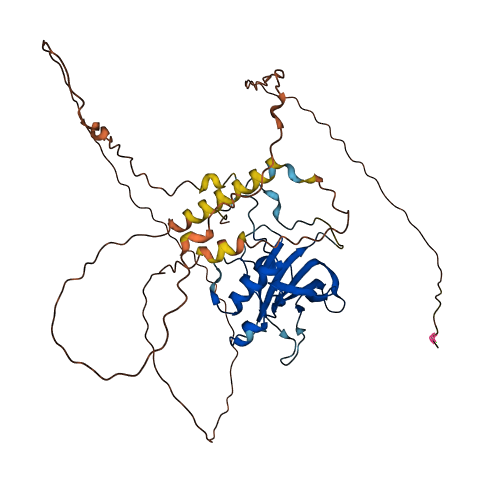
- 60.31
- Protein Structure from AlphaFold and UniProt
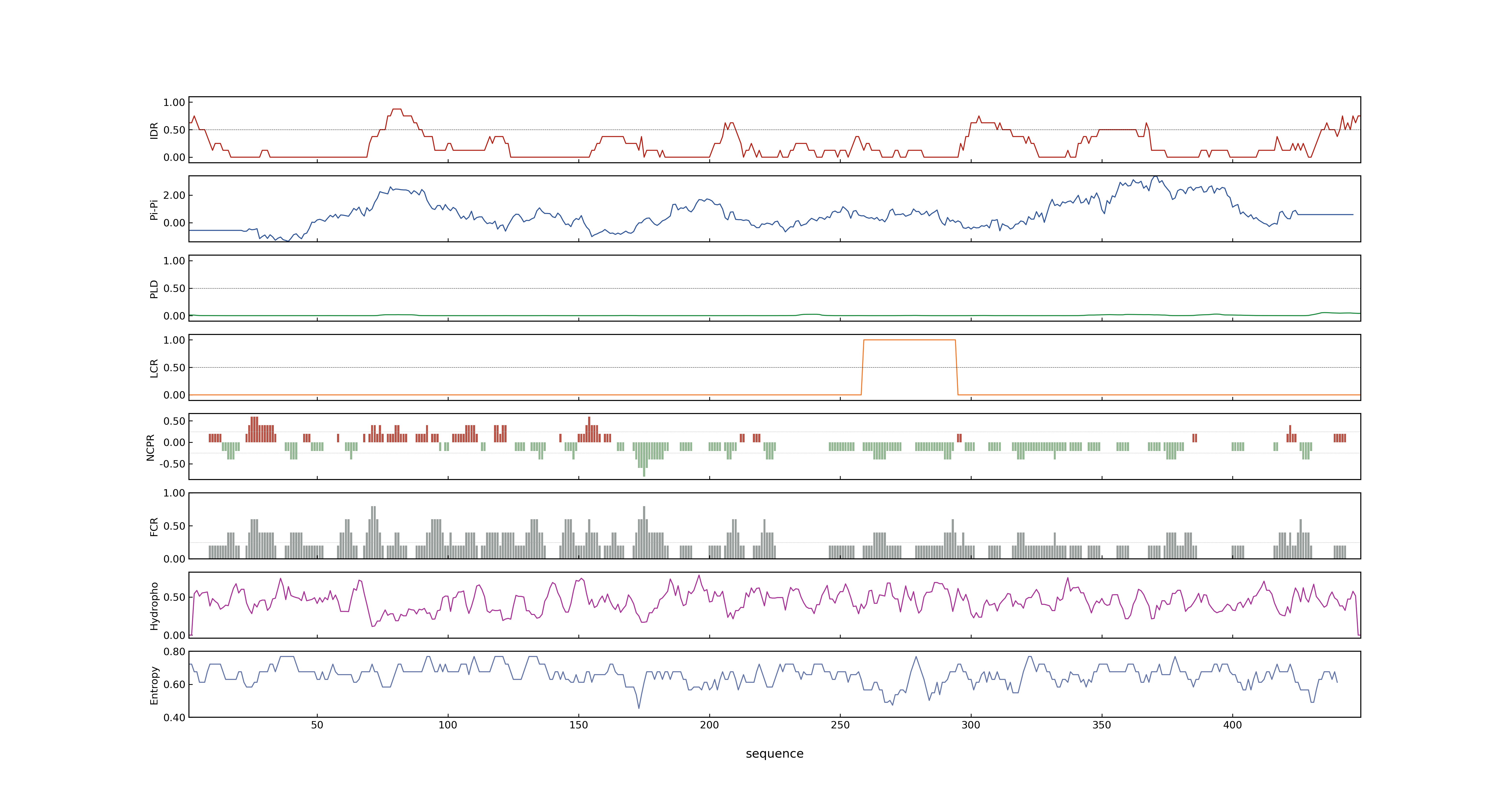
- MolPhase score
- LOC_Os05g35170.1: 0.63675153
- LOC_Os05g35170.2: 0.63675153
- LOC_Os05g35170.3: 0.69069832
- LOC_Os05g35170.4: 0.02431789
- LOC_Os05g35170.5: 0.05193086
- MolPhase Result
- Publication
- Genbank accession number
- Key message
- We identified a novel transcription factor of rice and barley, IDEF2, which specifically binds to the iron deficiency-responsive cis-acting element 2 (IDE2) by yeast one-hybrid screening
- IDEF2 belongs to an uncharacterized branch of the NAC transcription factor family and exhibits novel properties of sequence recognition
- A novel NAC transcription factor, IDEF2, that recognizes the iron deficiency-responsive element 2 regulates the genes involved in iron homeostasis in plants
- IDEF2 transcripts are constitutively present in rice roots and leaves
- IDEF2 was expressed in pollen, ovaries and the dorsal vascular region of the endosperm
- During seed germination, IDEF1 and IDEF2 were expressed in the endosperm and embryo
- IDEF1 and IDEF2 expression was dominant in leaf mesophyll and vascular cells, respectively
- The spatial expression and regulation of transcription factors IDEF1 and IDEF2
- KEY RESULTS: IDEF1 and IDEF2 were highly expressed in the basal parts of the lateral roots and vascular bundles
- The iron deficiency-responsive cis-acting element binding factors 1 and 2 (IDEF1 and IDEF2) regulate transcriptional response to iron deficiency in rice roots
- The spatial expression patterns and gene regulation of IDEF1 and IDEF2 in roots are generally conserved under conditions of iron sufficiency and deficiency, suggesting complicated interactions with unknown factors for sensing and transmitting iron-deficiency signals
- CONCLUSIONS: IDEF1 and IDEF2 are constitutively expressed during both vegetative and reproductive stages
- Several genes up-regulated by iron deficiency, including the Fe(II)-nicotianamine transporter gene OsYSL2, were less induced by iron deficiency in the RNAi rice of IDEF2, suggesting that IDEF2 is involved in the regulation of these genes
- Expression patterns of the target genes of IDEF1 and IDEF2 were analysed using transformants with induced or repressed expression of IDEF1 or IDEF2 grown in iron-rich or in iron-deficient solutions for 1 d
- In addition, the expression patterns of IDEF2 target genes were similar between iron-rich conditions and early or subsequent iron deficiency
- Repression of the function of IDEF2 by the RNA interference (RNAi) technique and chimeric repressor gene-silencing technology (CRES-T) caused aberrant iron homeostasis in rice
- Connection
- IDEF2, OsYSL2, A novel NAC transcription factor, IDEF2, that recognizes the iron deficiency-responsive element 2 regulates the genes involved in iron homeostasis in plants, Several genes up-regulated by iron deficiency, including the Fe(II)-nicotianamine transporter gene OsYSL2, were less induced by iron deficiency in the RNAi rice of IDEF2, suggesting that IDEF2 is involved in the regulation of these genes
- IDEF2, OsYSL2, A novel NAC transcription factor, IDEF2, that recognizes the iron deficiency-responsive element 2 regulates the genes involved in iron homeostasis in plants, IDEF2 bound to OsYSL2 promoter region containing the binding core site, suggesting direct regulation of OsYSL2 expression
- IDEF1, IDEF2, The spatial expression and regulation of transcription factors IDEF1 and IDEF2, The iron deficiency-responsive cis-acting element binding factors 1 and 2 (IDEF1 and IDEF2) regulate transcriptional response to iron deficiency in rice roots
- IDEF1, IDEF2, The spatial expression and regulation of transcription factors IDEF1 and IDEF2, Clarification of the functions of IDEF1 and IDEF2 could uncover the gene regulation mechanism
- IDEF1, IDEF2, The spatial expression and regulation of transcription factors IDEF1 and IDEF2, METHODS: Spatial patterns of IDEF1 and IDEF2 expression were analysed by histochemical staining of IDEF1 and IDEF2 promoter-GUS transgenic rice lines
- IDEF1, IDEF2, The spatial expression and regulation of transcription factors IDEF1 and IDEF2, Expression patterns of the target genes of IDEF1 and IDEF2 were analysed using transformants with induced or repressed expression of IDEF1 or IDEF2 grown in iron-rich or in iron-deficient solutions for 1 d
- IDEF1, IDEF2, The spatial expression and regulation of transcription factors IDEF1 and IDEF2, KEY RESULTS: IDEF1 and IDEF2 were highly expressed in the basal parts of the lateral roots and vascular bundles
- IDEF1, IDEF2, The spatial expression and regulation of transcription factors IDEF1 and IDEF2, IDEF1 and IDEF2 expression was dominant in leaf mesophyll and vascular cells, respectively
- IDEF1, IDEF2, The spatial expression and regulation of transcription factors IDEF1 and IDEF2, During seed germination, IDEF1 and IDEF2 were expressed in the endosperm and embryo
- IDEF1, IDEF2, The spatial expression and regulation of transcription factors IDEF1 and IDEF2, CONCLUSIONS: IDEF1 and IDEF2 are constitutively expressed during both vegetative and reproductive stages
- IDEF1, IDEF2, The spatial expression and regulation of transcription factors IDEF1 and IDEF2, The spatial expression patterns of IDEF1 and IDEF2 overlap with their target genes in restricted cell types, but not in all cells
- IDEF1, IDEF2, The spatial expression and regulation of transcription factors IDEF1 and IDEF2, The spatial expression patterns and gene regulation of IDEF1 and IDEF2 in roots are generally conserved under conditions of iron sufficiency and deficiency, suggesting complicated interactions with unknown factors for sensing and transmitting iron-deficiency signals
- IDEF1, IDEF2, The spatial expression and regulation of transcription factors IDEF1 and IDEF2, The spatial expression and regulation of transcription factors IDEF1 and IDEF2
Prev Next