- Information
- Symbol: LMM5.1,SPL33
- MSU: LOC_Os01g02720
- RAPdb: Os01g0116600
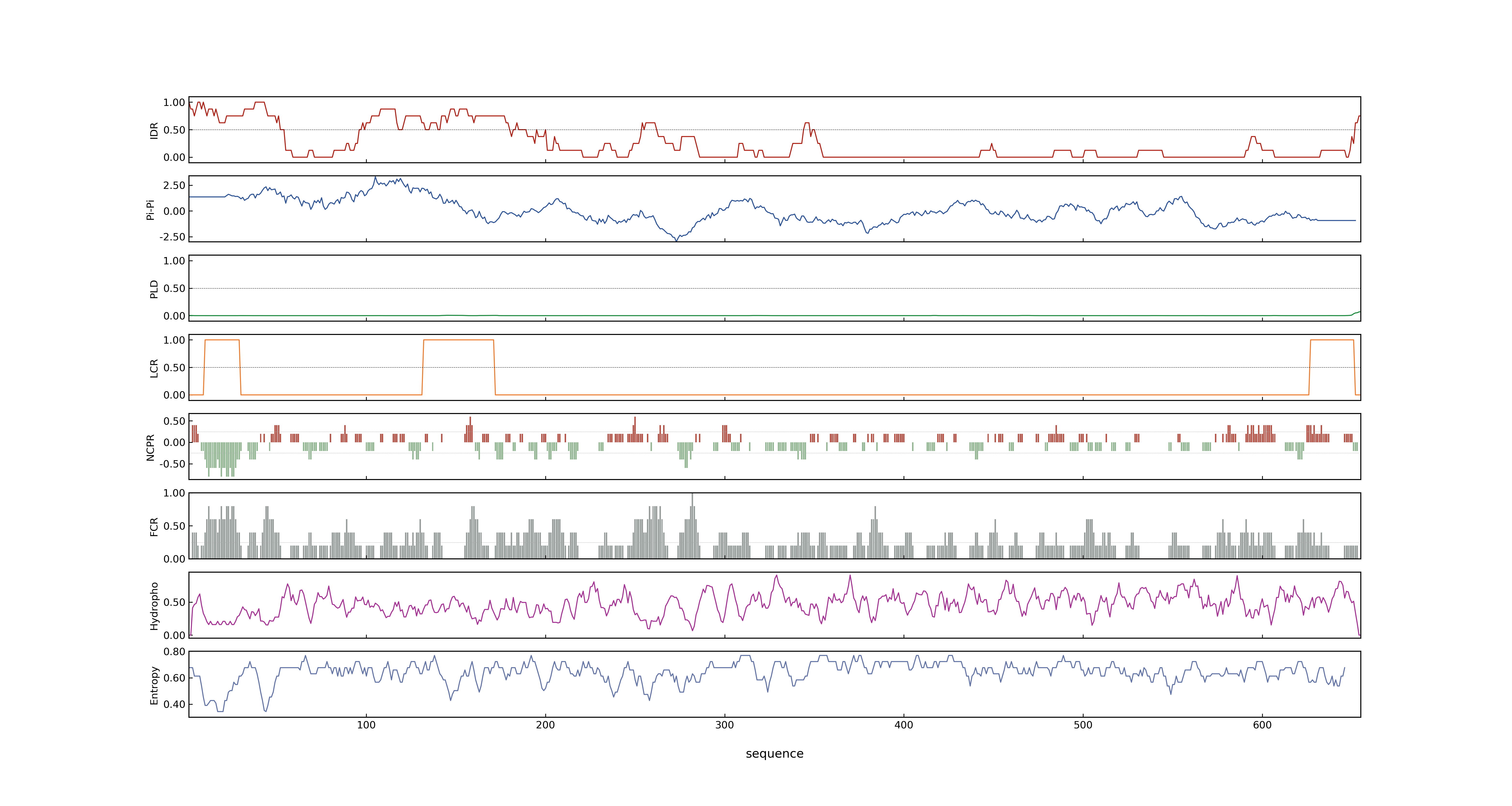
- PSP score
- LOC_Os01g02720.1: 0.868
- LOC_Os01g02720.2: 0.868
- PLAAC score
- LOC_Os01g02720.1: 0
- LOC_Os01g02720.2: 0
- pLDDT score
- NA
- MolPhase score
- LOC_Os01g02720.1: 0.99554258
- LOC_Os01g02720.2: 0.99554258
- MolPhase Result
- Publication
-
Genbank accession number
- Key message
- LMM5.1 and LMM5.4, two eukaryotic translation elongation factor 1A-like gene family members, negatively affect cell death and disease resistance in rice.
- spl33 exhibited programmed cell death-mediated cell death and early leaf senescence, as evidenced by analyses of four histochemical markers, namely H2O2 accumulation, cell death, callose accumulation and TUNEL-positive nuclei, and by four indicators, namely loss of chlorophyll, breakdown of chloroplasts, down-regulation of photosynthesis-related genes, and up-regulation of senescence-associated genes
- Transcriptome analysis of the spl33 mutant and its wild type provided further evidence for the biological effects of loss of SPL33 function in cell death, leaf senescence and defense responses in rice
- Defense responses were induced in the spl33 mutant, as shown by enhanced resistance to both the fungal pathogen Magnaporthe oryzae and the bacterial pathogen Xanthomonas oryzae pv
- Detailed analyses showed that reactive oxygen species accumulation may be the cause of cell death in the spl33 mutant, whereas uncontrolled activation of multiple innate immunity-related receptor genes and signaling molecules may be responsible for the enhanced disease resistance observed in spl33
- SPL33 encodes a eukaryotic translation elongation factor 1 alpha (eEF1A)-like protein consisting of a non-functional zinc finger domain and three functional EF-Tu domains
- Connection
Prev Next