- Information
- Symbol: MOC1
- MSU: LOC_Os06g40780
- RAPdb: Os06g0610300,Os06g0610350
- PSP score
- LOC_Os06g40780.1: 0.9393
- PLAAC score
- LOC_Os06g40780.1: 0
- pLDDT score
- 80.81
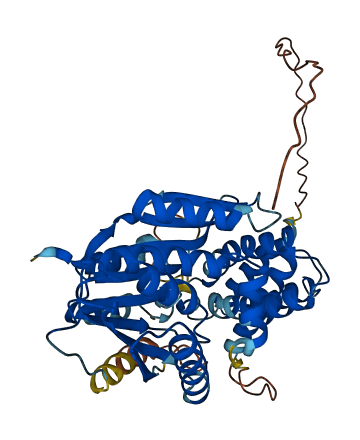
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os06g40780.1: 0.99819642
- MolPhase Result
- Publication
- Identification and functional analysis of the MOC1 interacting protein 1, 2010, Journal of Genetics and Genomics.
- A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice, 2011, Mol Plant.
- Control of tillering in rice, 2003, Nature.
- Rice APC/CTE controls tillering by mediating the degradation of MONOCULM 1, 2012, Nat Commun.
- Degradation of MONOCULM 1 by APC/CTAD1 regulates rice tillering, 2012, Nat Commun.
- Comparative sequence analysis of MONOCULM1-orthologous regions in 14 Oryza genomes, 2009, Proc Natl Acad Sci U S A.
- Novel function of a putative MOC1 ortholog associated with spikelet number per spike in common wheat., 2015, Sci Rep.
- Genbank accession number
- Key message
- Here we report the isolation and characterization of MONOCULM 1 (MOC1), a gene that is important in the control of rice tillering
- The moc1 mutant plants have only a main culm without any tillers owing to a defect in the formation of tiller buds
- A large genomic region surrounding the MONOCULM1 (MOC1) locus was chosen for study in 14 Oryza species, including 10 diploids and 4 allotetraploids
- Ghd8 up-regulated MOC1, a key gene controlling tillering and branching; this increased the number of tillers, primary and secondary branches, thus producing 50% more grains per plant
- To further elucidate the molecular mechanism of MOC1 involved in the regulation of rice tillering, we performed a yeast-two-hybrid screening to identify MOC1 interacting proteins (MIPs)
- In-depth characterization of the context of MIP1 and MOC1 would further our understanding of molecular regulatory mechanisms of rice tillering
- We show that TE coexpresses with MOC1 in the axil of leaves, where the APC/C(TE) complex mediates the degradation of MOC1 by the ubiquitin-26S proteasome pathway, and consequently downregulates the expression of the meristem identity gene Oryza sativa homeobox 1, thus repressing axillary meristem initiation and formation
- The MONOCULM 1 (MOC1) gene is the first identified key regulator controlling rice tiller number; however, the underlying mechanism remains to be elucidated
- Rice MONOCULM 1 (MOC1) and its orthologues LS/LAS (lateral suppressor in tomato and Arabidopsis) are key promoting factors of shoot branching and tillering in higher plants
- MOC1 encodes a putative GRAS family nuclear protein that is expressed mainly in the axillary buds and functions to initiate axillary buds and to promote their outgrowth
- Our previous study has demonstrated that the MONOCULM1 (MOC1) gene is a key component that controls the formation of rice tiller buds
- Novel function of a putative MOC1 ortholog associated with spikelet number per spike in common wheat.
- Tomato Ls, Arabidopsis LAS and rice MOC1 are orthologous genes regulating axillary meristem initiation and outgrowth
- Connection
- MOC1, OsHUB1~MIP1, Identification and functional analysis of the MOC1 interacting protein 1, Here we reported that MIP1 interacted with MOC1 both in vitro and in vivo
- MOC1, OsHUB1~MIP1, Identification and functional analysis of the MOC1 interacting protein 1, In-depth characterization of the context of MIP1 and MOC1 would further our understanding of molecular regulatory mechanisms of rice tillering
- Hd5~DTH8~Ghd8~OsHAP3H~LHD1~EF8~CAR8~OsNF-YB11, MOC1, A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice, Ghd8 up-regulated MOC1, a key gene controlling tillering and branching; this increased the number of tillers, primary and secondary branches, thus producing 50% more grains per plant
- MOC1, OsCCS52A~TAD~TE, Rice APC/CTE controls tillering by mediating the degradation of MONOCULM 1, TE physically interacts with MOC1 and OsCDC27
Prev Next