- Information
- Symbol: ONAC122,OsNAC10
- MSU: LOC_Os11g03300
- RAPdb: Os11g0126900
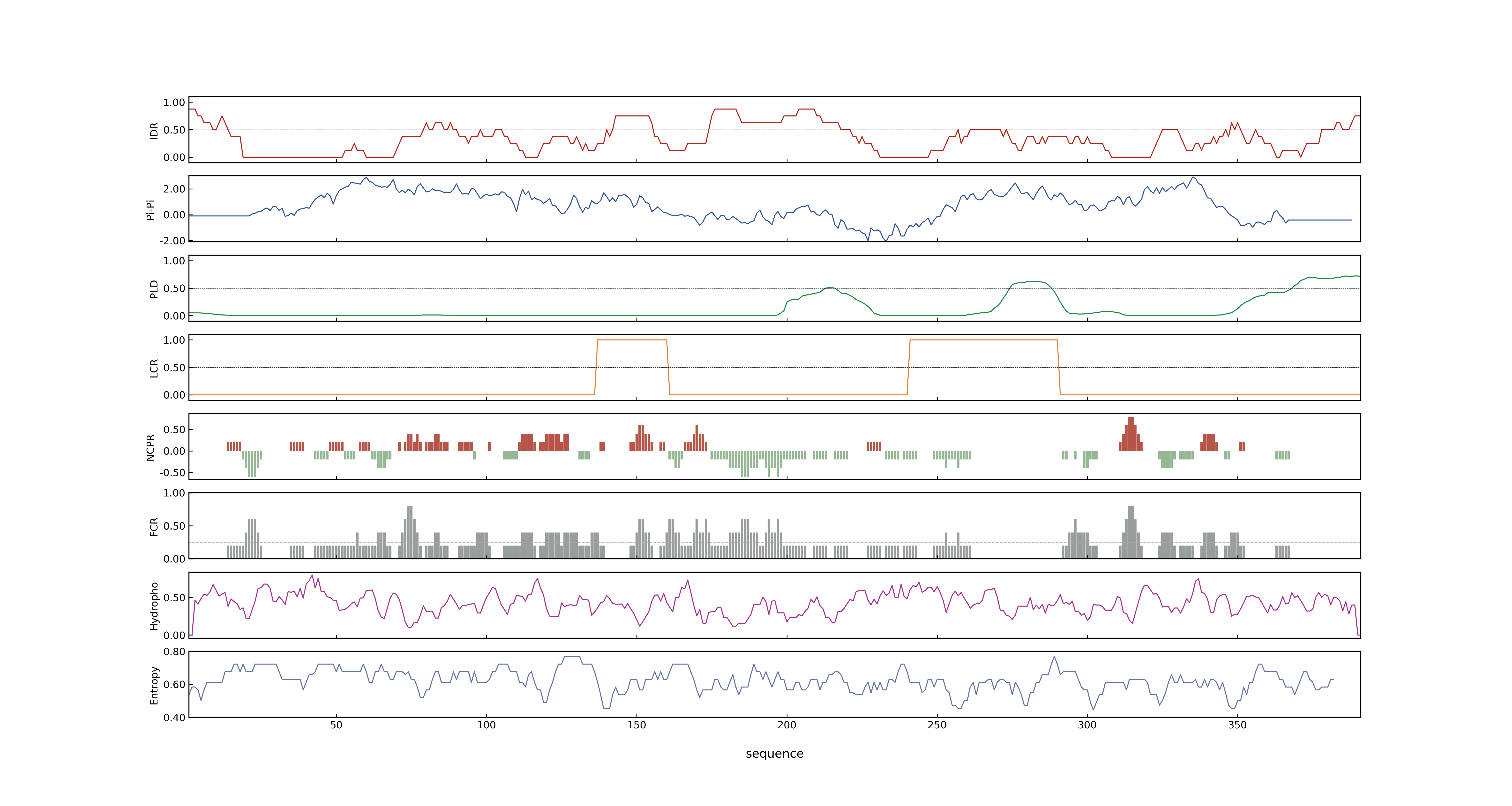
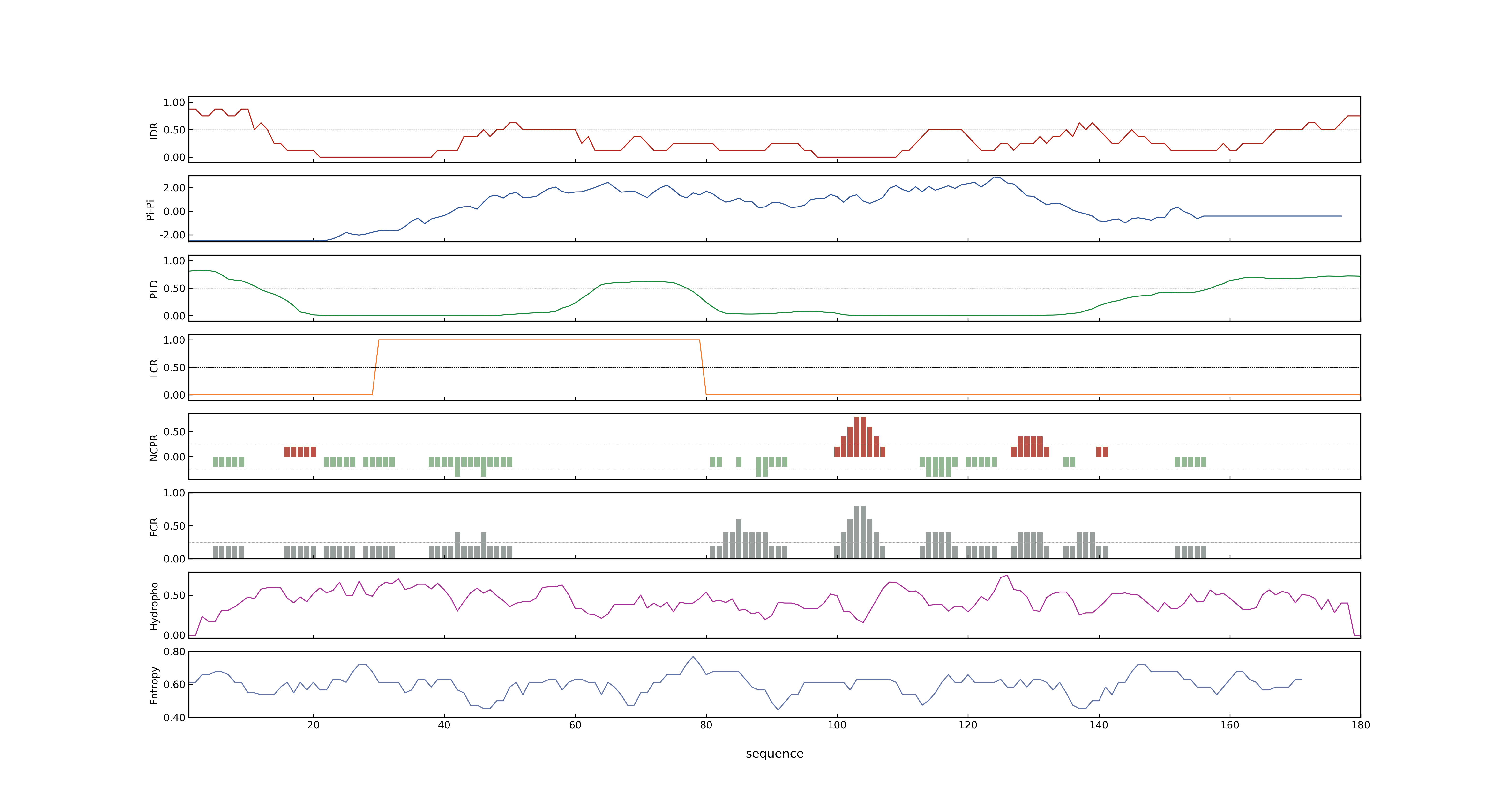
- PSP score
- LOC_Os11g03300.1: 0.9435
- LOC_Os11g03300.2: 0.4428
- PLAAC score
- LOC_Os11g03300.1: 0
- LOC_Os11g03300.2: 0
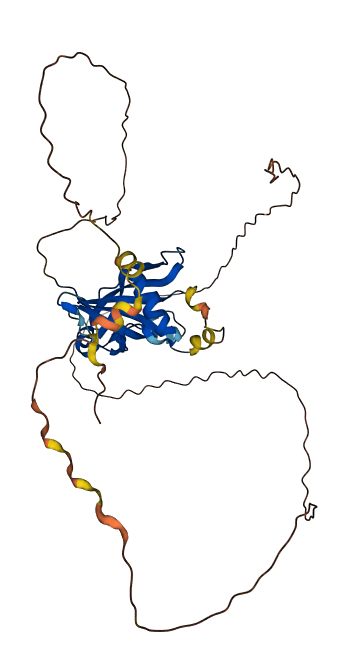
- pLDDT score
- 60.31
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os11g03300.1: 0.99983489
- LOC_Os11g03300.2: 0.95885497
- MolPhase Result
- Publication
- Genbank accession number
- Key message
- More importantly, the RCc3:OsNAC10 plants showed significantly enhanced drought tolerance at the reproductive stage, increasing grain yield by 25% to 42% and by 5% to 14% over controls in the field under drought and normal conditions, respectively
- ONAC122 and ONAC131 were also induced by treatment with salicylic acid, methyl jasmonate or 1-aminocyclopropane-1-carboxylic acid (a precursor of ethylene)
- Our results suggest that both ONAC122 and ONAC131 have important roles in rice disease resistance responses through the regulated expression of other defense- and signaling-related genes
- Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea
- Here, we characterized two rice pathogen-responsive NAC transcription factors, ONAC122 and ONAC131
- Grain yield of GOS2:OsNAC10 plants in the field, in contrast, remained similar to that of controls under both normal and drought conditions
- Overall, our results demonstrated that root-specific overexpression of OsNAC10 enlarges roots, enhancing drought tolerance of transgenic plants, which increases grain yield significantly under field drought conditions
- Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions
- Overexpression of OsNAC10 in rice under the control of the constitutive promoter GOS2 and the root-specific promoter RCc3 increased the plant tolerance to drought, high salinity, and low temperature at the vegetative stage
- Here, we report the results of a functional genomics approach that identified a rice NAC (an acronym for NAM [No Apical Meristem], ATAF1-2, and CUC2 [Cup-Shaped Cotyledon]) domain gene, OsNAC10, which improved performance of transgenic rice plants under field drought conditions
- OsNAC10, one of the effective members selected from prescreening, is expressed predominantly in roots and panicles and induced by drought, high salinity, and abscisic acid
- ONAC122 and ONAC131 expression was induced after infection by Magnaporthe grisea, the causal agent of rice blast disease, and the M
- These differences in performance under field drought conditions reflect the differences in expression of OsNAC10-dependent target genes in roots as well as in leaves of the two transgenic plants, as revealed by microarray analyses
- Root diameter of the RCc3:OsNAC10 plants was thicker by 1
- Connection
- ONAC122~OsNAC10, OsWRKY45~WRKY45, Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea, OsLOX, OsPR1a, OsWRKY45 and OsNH1) were down-regulated in plants silenced for ONAC122 or ONAC131 expression via the BMV-based silencing system
- ONAC122~OsNAC10, ONAC131, Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea, Here, we characterized two rice pathogen-responsive NAC transcription factors, ONAC122 and ONAC131
- ONAC122~OsNAC10, ONAC131, Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea, ONAC122 and ONAC131 expression was induced after infection by Magnaporthe grisea, the causal agent of rice blast disease, and the M
- ONAC122~OsNAC10, ONAC131, Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea, ONAC122 and ONAC131 were also induced by treatment with salicylic acid, methyl jasmonate or 1-aminocyclopropane-1-carboxylic acid (a precursor of ethylene)
- ONAC122~OsNAC10, ONAC131, Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea, Silencing ONAC122 or ONAC131 expression using a newly modified Brome mosaic virus (BMV)-based silencing vector resulted in an enhanced susceptibility to M
- ONAC122~OsNAC10, ONAC131, Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea, OsLOX, OsPR1a, OsWRKY45 and OsNH1) were down-regulated in plants silenced for ONAC122 or ONAC131 expression via the BMV-based silencing system
- ONAC122~OsNAC10, ONAC131, Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea, Our results suggest that both ONAC122 and ONAC131 have important roles in rice disease resistance responses through the regulated expression of other defense- and signaling-related genes
- ONAC122~OsNAC10, ONAC131, Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea, Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea
- ONAC122~OsNAC10, OsNPR1~NH1, Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea, OsLOX, OsPR1a, OsWRKY45 and OsNH1) were down-regulated in plants silenced for ONAC122 or ONAC131 expression via the BMV-based silencing system
- ONAC122~OsNAC10, OsPR1a~OsSCP, Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea, OsLOX, OsPR1a, OsWRKY45 and OsNH1) were down-regulated in plants silenced for ONAC122 or ONAC131 expression via the BMV-based silencing system
Prev Next