- Information
- Symbol: OSH71,Oskn2
- MSU: LOC_Os05g03884
- RAPdb: Os05g0129700
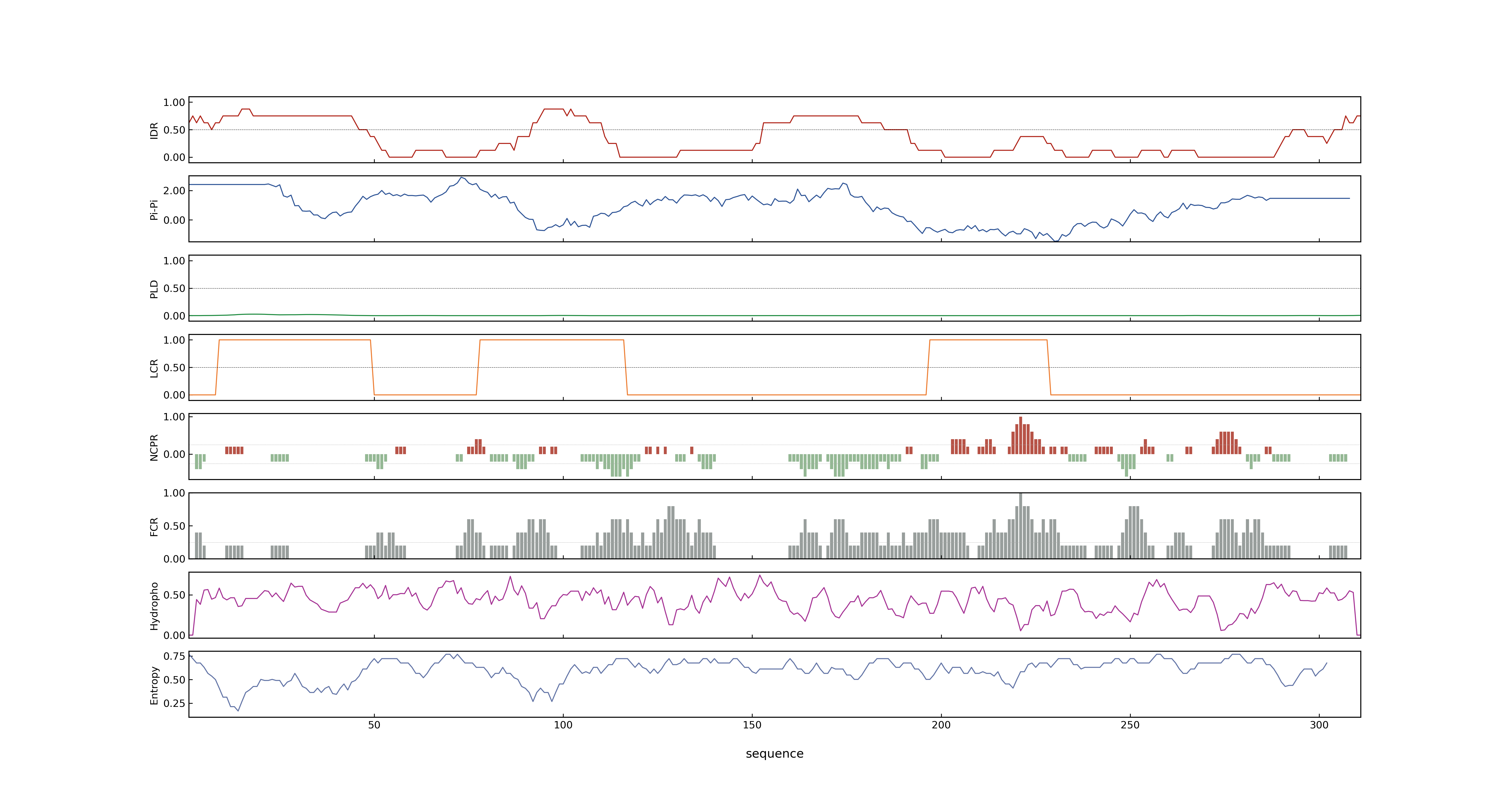
- PSP score
- LOC_Os05g03884.1: 0.2271
- PLAAC score
- LOC_Os05g03884.1: 0
- pLDDT score
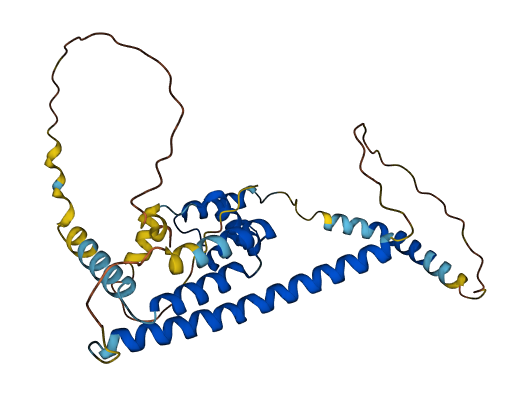
- 73.32
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os05g03884.1: 0.99911659
- MolPhase Result
- Publication
- Overexpression of rice OSH genes induces ectopic shoots on leaf sheaths of transgenic rice plants, 2000, Dev Biol.
- Disruption of KNOX gene suppression in leaf by introducing its cDNA in rice, 2008, Plant Science.
- Characterization of the KNOX class homeobox genes Oskn2 and Oskn3 identified in a collection of cDNA libraries covering the early stages of rice embryogenesis, 1999, Plant Mol Biol.
- Developmental regulation and downstream effects of the knox class homeobox genes Oskn2 and Oskn3 from rice, 2002, Plant Mol Biol.
- KNOX homeobox genes are sufficient in maintaining cultured cells in an undifferentiated state in rice, 2001, Genesis.
- Regional expression of the rice KN1-type homeobox gene family during embryo, shoot, and flower development, 1999, Plant Cell.
- Genbank accession number
- Key message
- Whereas Oskn3 transformants showed the most pronounced phenotypic effects during vegetative development, Oskn2 transformants showed relatively mild alterations in the vegetative phase but a more severely affected flower morphology
- Ectopic expression of Oskn2 or Oskn3 induced similar defects in panicle branching
- We produced transgenic rice calli, which constitutively express each of four KNOX family class 1 homeobox genes of rice, OSH1, OSH16, OSH15, and OSH71, and found that constitutive and ectopic expression of such genes inhibits normal regeneration from transformed calli, which showed continuous growth around their shoot-regenerating stages
- Connection
- OSH1~Oskn1, OSH71~Oskn2, Characterization of the KNOX class homeobox genes Oskn2 and Oskn3 identified in a collection of cDNA libraries covering the early stages of rice embryogenesis, Two of these, Oskn2 and Oskn3, encode newly described KNOX genes, whereas the third (Oskn1) corresponds to the previously described OSH1 gene
- OSH1~Oskn1, OSH71~Oskn2, Characterization of the KNOX class homeobox genes Oskn2 and Oskn3 identified in a collection of cDNA libraries covering the early stages of rice embryogenesis, Transgenic expression of Oskn2 and Oskn3 in tobacco further supported their involvement in cell fate determination, like previously reported for Knotted1 and OSH1 ectopic expression
- D6~OSH15~Oskn3, OSH71~Oskn2, Characterization of the KNOX class homeobox genes Oskn2 and Oskn3 identified in a collection of cDNA libraries covering the early stages of rice embryogenesis, Two of these, Oskn2 and Oskn3, encode newly described KNOX genes, whereas the third (Oskn1) corresponds to the previously described OSH1 gene
- D6~OSH15~Oskn3, OSH71~Oskn2, Characterization of the KNOX class homeobox genes Oskn2 and Oskn3 identified in a collection of cDNA libraries covering the early stages of rice embryogenesis, Transgenic expression of Oskn2 and Oskn3 in tobacco further supported their involvement in cell fate determination, like previously reported for Knotted1 and OSH1 ectopic expression
- D6~OSH15~Oskn3, OSH71~Oskn2, Characterization of the KNOX class homeobox genes Oskn2 and Oskn3 identified in a collection of cDNA libraries covering the early stages of rice embryogenesis, Whereas Oskn3 transformants showed the most pronounced phenotypic effects during vegetative development, Oskn2 transformants showed relatively mild alterations in the vegetative phase but a more severely affected flower morphology
- D6~OSH15~Oskn3, OSH71~Oskn2, Characterization of the KNOX class homeobox genes Oskn2 and Oskn3 identified in a collection of cDNA libraries covering the early stages of rice embryogenesis, Characterization of the KNOX class homeobox genes Oskn2 and Oskn3 identified in a collection of cDNA libraries covering the early stages of rice embryogenesis
- D6~OSH15~Oskn3, OSH71~Oskn2, Developmental regulation and downstream effects of the knox class homeobox genes Oskn2 and Oskn3 from rice, Here, the transcriptional control of two rice knox genes, Oskn2 and Oskn3, was addressed, showing that the promoter sequences of both genes mediate the initial down-regulation during lateral organ formation, but are insufficient to keep expression in lateral organs stably off
- D6~OSH15~Oskn3, OSH71~Oskn2, Developmental regulation and downstream effects of the knox class homeobox genes Oskn2 and Oskn3 from rice, Ectopic expression of Oskn2 or Oskn3 induced similar defects in panicle branching
- D6~OSH15~Oskn3, OSH71~Oskn2, Developmental regulation and downstream effects of the knox class homeobox genes Oskn2 and Oskn3 from rice, This was supported by the observation that Oskn3 protein but not Oskn2 could interact with two reported recognition sequences of a KNOX protein from barley
- D6~OSH15~Oskn3, OSH71~Oskn2, Developmental regulation and downstream effects of the knox class homeobox genes Oskn2 and Oskn3 from rice, Finally, protein-protein interactions may contribute to the functioning of KNOX proteins, as the ability of Oskn3 and Oskn2 to form heterodimers could be demonstrated
- D6~OSH15~Oskn3, OSH71~Oskn2, Developmental regulation and downstream effects of the knox class homeobox genes Oskn2 and Oskn3 from rice, Developmental regulation and downstream effects of the knox class homeobox genes Oskn2 and Oskn3 from rice
- OSH1~Oskn1, OSH71~Oskn2, KNOX homeobox genes are sufficient in maintaining cultured cells in an undifferentiated state in rice, We produced transgenic rice calli, which constitutively express each of four KNOX family class 1 homeobox genes of rice, OSH1, OSH16, OSH15, and OSH71, and found that constitutive and ectopic expression of such genes inhibits normal regeneration from transformed calli, which showed continuous growth around their shoot-regenerating stages
- D6~OSH15~Oskn3, OSH71~Oskn2, KNOX homeobox genes are sufficient in maintaining cultured cells in an undifferentiated state in rice, We produced transgenic rice calli, which constitutively express each of four KNOX family class 1 homeobox genes of rice, OSH1, OSH16, OSH15, and OSH71, and found that constitutive and ectopic expression of such genes inhibits normal regeneration from transformed calli, which showed continuous growth around their shoot-regenerating stages
- OsARID3, OSH71~Oskn2, OsARID3, an AT-rich Interaction Domain-containing protein, is required for shoot meristem development in rice., Furthermore, our chromatin immunoprecipitation results demonstrate that OsARID3 binds directly to the KNOXI gene OSH71, the auxin biosynthetic genes OsYUC1 and OsYUC6, and the cytokinin biosynthetic genes OsIPT2 and OsIPT7
- OSH71~Oskn2, OsIPT2, OsARID3, an AT-rich Interaction Domain-containing protein, is required for shoot meristem development in rice., Furthermore, our chromatin immunoprecipitation results demonstrate that OsARID3 binds directly to the KNOXI gene OSH71, the auxin biosynthetic genes OsYUC1 and OsYUC6, and the cytokinin biosynthetic genes OsIPT2 and OsIPT7
- OSH71~Oskn2, OsIPT7, OsARID3, an AT-rich Interaction Domain-containing protein, is required for shoot meristem development in rice., Furthermore, our chromatin immunoprecipitation results demonstrate that OsARID3 binds directly to the KNOXI gene OSH71, the auxin biosynthetic genes OsYUC1 and OsYUC6, and the cytokinin biosynthetic genes OsIPT2 and OsIPT7
- OsGRF5, OSH71~Oskn2, Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors., Conversely, RNA interference silencing of OsGRF3, OsGRF4, and OsGRF5 resulted in dwarfism, delayed growth and inflorescence formation, and up-regulation of Oskn2
- OsGRF3, OSH71~Oskn2, Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors., A yeast (Saccharomyces cerevisiae) one-hybrid screen with the upstream sequence of the KNOX gene Oskn2 from rice (Oryza sativa) resulted in isolation of OsGRF3 and OsGRF10
- OsGRF3, OSH71~Oskn2, Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors., These effects were associated with down-regulation of endogenous Oskn2 expression by OsGRF3
- OsGRF3, OSH71~Oskn2, Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors., Conversely, RNA interference silencing of OsGRF3, OsGRF4, and OsGRF5 resulted in dwarfism, delayed growth and inflorescence formation, and up-regulation of Oskn2
- GS2~OsGRF4~GL2~GLW2, OSH71~Oskn2, Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors., Conversely, RNA interference silencing of OsGRF3, OsGRF4, and OsGRF5 resulted in dwarfism, delayed growth and inflorescence formation, and up-regulation of Oskn2
- OsGRF10, OSH71~Oskn2, Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors., A yeast (Saccharomyces cerevisiae) one-hybrid screen with the upstream sequence of the KNOX gene Oskn2 from rice (Oryza sativa) resulted in isolation of OsGRF3 and OsGRF10
- OsARID3, OSH71~Oskn2, OsARID3, an AT-rich Interaction Domain-containing protein, is required for shoot meristem development in rice., Furthermore, chromatin immunoprecipitation results demonstrate that OsARID3 binds directly to the KNOXI gene OSH71, the auxin biosynthetic genes OsYUC1 and OsYUC6, and the cytokinin biosynthetic genes OsIPT2 and OsIPT7
Prev Next