- Information
- Symbol: OSMADS3
- MSU: LOC_Os01g10504
- RAPdb: Os01g0201700
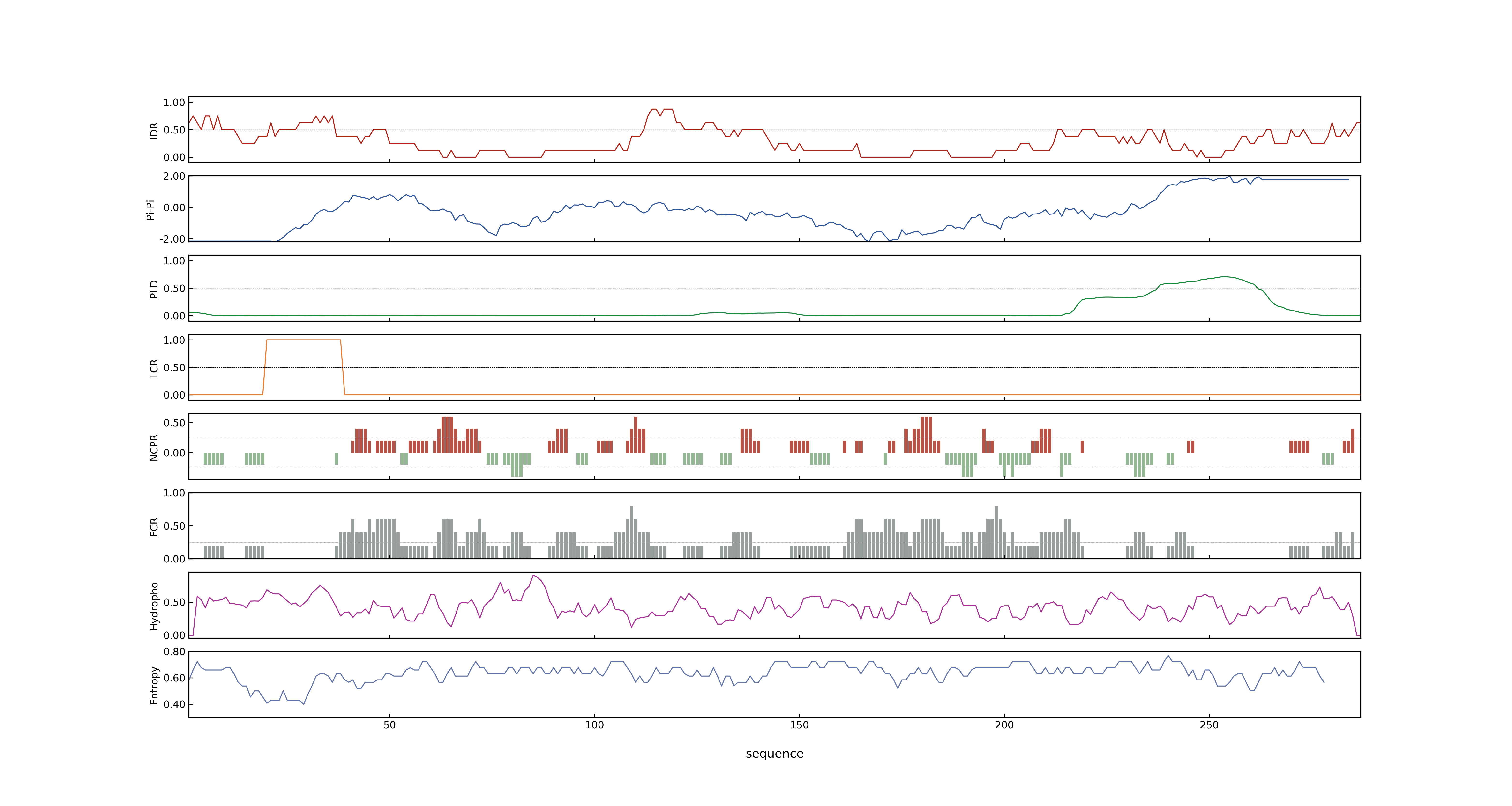
- PSP score
- LOC_Os01g10504.2: 0.0277
- LOC_Os01g10504.1: 0.1028
- LOC_Os01g10504.3: 0.0277
- PLAAC score
- LOC_Os01g10504.2: 0
- LOC_Os01g10504.1: 0
- LOC_Os01g10504.3: 0
- pLDDT score
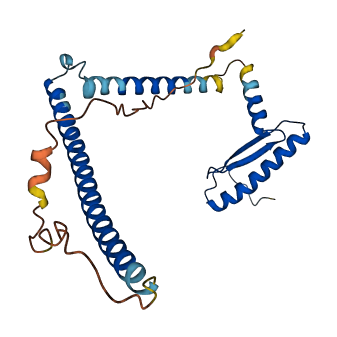
- 79.31
- Protein Structure from AlphaFold and UniProt
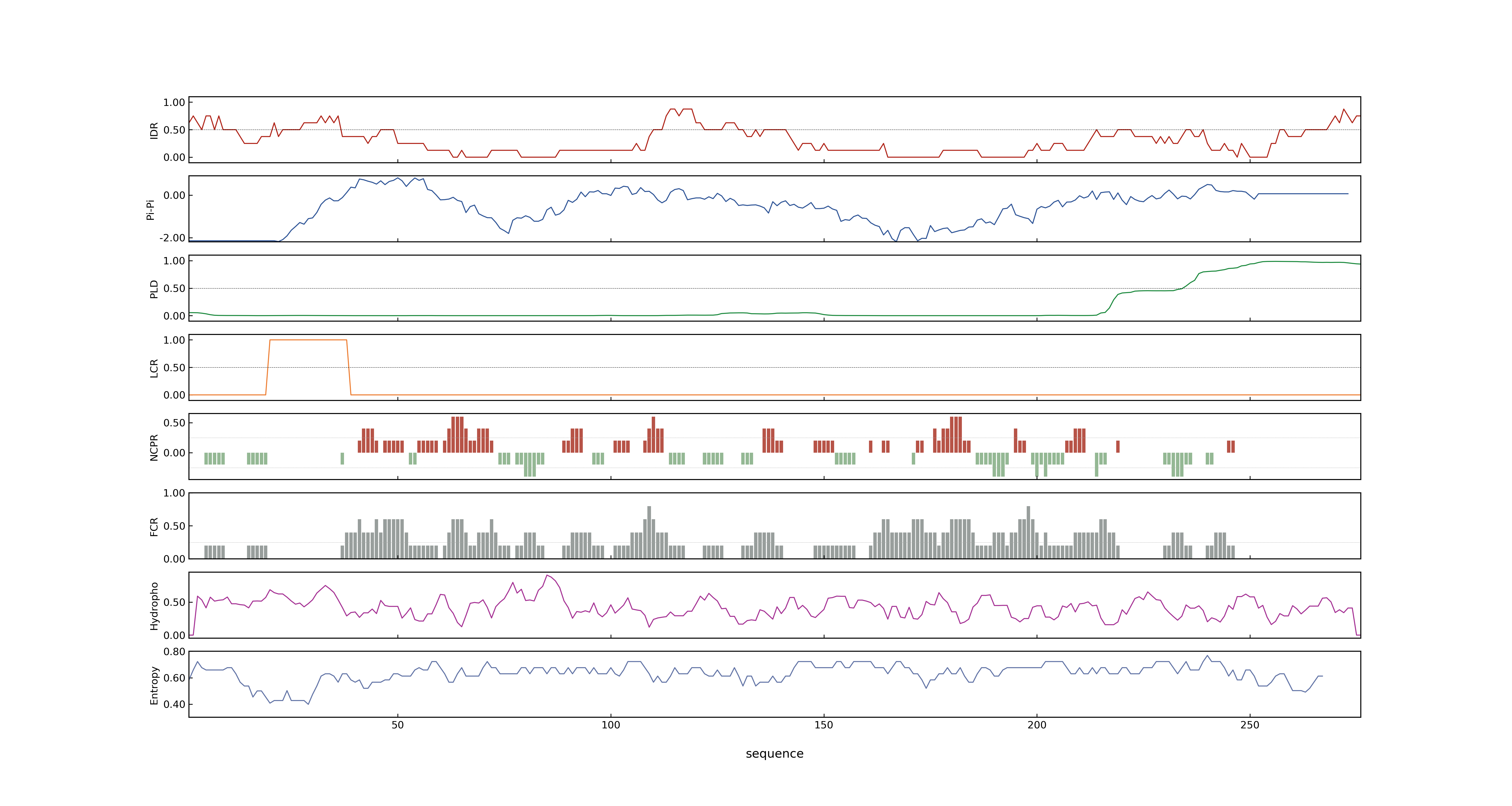
- MolPhase score
- LOC_Os01g10504.1: 0.97763305
- LOC_Os01g10504.2: 0.79743862
- LOC_Os01g10504.3: 0.79743862
- MolPhase Result
- Publication
- MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress, 2007, BMC Genomics.
- Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy, 2011, Plant Physiol.
- Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy, 2011, Plant Cell.
- Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa, 2006, Plant Cell.
- Rice MADS3 regulates ROS homeostasis during late anther development, 2011, Plant Cell.
- Rice MADS6 interacts with the floral homeotic genes SUPERWOMAN1, MADS3, MADS58, MADS13, and DROOPING LEAF in specifying floral organ identities and meristem fate, 2011, Plant Cell.
- Ectopic Expression of OsMADS3, a Rice Ortholog of AGAMOUS, Caused a Homeotic Transformation of Lodicules to Stamens in Transgenic Rice Plants, 2002, Plant and Cell Physiology.
- Loss of function of OsMADS3 via the insertion of a novel retrotransposon leads to recessive male sterility in rice Oryza sativa., 2015, Plant Sci.
- Genetic Enhancer Analysis Reveals that FLORAL ORGAN NUMBER2 and OsMADS3 Cooperatively Regulate Maintenance and Determinacy of the Flower Meristem in Rice., 2017, Plant Cell Physiol.
- Genbank accession number
- Key message
- A knockout line of OSMADS3, in which the gene is disrupted by T-DNA insertion, shows homeotic transformation of stamens into lodicules and ectopic development of lodicules in the second whorl near the palea where lodicules do not form in the wild type but carpels develop almost normally
- osmads13-3 osmads3-4 displayed a loss of floral meristem determinacy and generated abundant carpelloid structures containing severe defective ovules in the flower center, which were not detectable in the single mutant
- Collectively, we propose a model to illustrate the role of OsMADS3, DL, and OsMADS13 in the specification of flower organ identity and meristem determinacy in rice
- Here, we report the interactions of rice (Oryza sativa) floral homeotic genes, OsMADS3 (a C-class gene), OsMADS13 (a D-class gene), and DROOPING LEAF (DL), in specifying floral organ identities and floral meristem determinacy
- This indicates that OsMADS3 plays a synergistic role with OsMADS13 in both ovule development and floral meristem termination
- Strikingly, osmads3-4 dl-sup6 displayed a severe loss of floral meristem determinacy and produced supernumerary whorls of lodicule-like organs at the forth whorl, suggesting that OsMADS3 and DL synergistically terminate the floral meristem
- Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy
- As a consequence of the ectopic expression of the OsMADS3, lodicules were homeotically transformed into stamens
- Ectopic Expression of OsMADS3, a Rice Ortholog of AGAMOUS, Caused a Homeotic Transformation of Lodicules to Stamens in Transgenic Rice Plants
- In order to clarify the evolutionary relationship of floral organs between grasses and dicots, we expressed OsMADS3, a rice (Oryza sativa L
- Loss of function of OsMADS3 via the insertion of a novel retrotransposon leads to recessive male sterility in rice (Oryza sativa).
- Genetic Enhancer Analysis Reveals that FLORAL ORGAN NUMBER2 and OsMADS3 Cooperatively Regulate Maintenance and Determinacy of the Flower Meristem in Rice.
- Disruption of OsMADS3 in the fon2 mutant by CRISPR-Cas9 technology caused a flower phenotype similar to that of 2B-424, confirming that the gene responsible for enhancement of fon2 was OsMADS3
- These findings suggest that OsMADS3 is involved not only in FM determinacy in late flower development but also in FM activity in early flower development
- Morphological analysis showed that the fon2 and osmads3 mutations synergistically affected pistil development and FM determinacy
- Connection
- DL, OSMADS3, Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy, Here, we report the interactions of rice (Oryza sativa) floral homeotic genes, OsMADS3 (a C-class gene), OsMADS13 (a D-class gene), and DROOPING LEAF (DL), in specifying floral organ identities and floral meristem determinacy
- DL, OSMADS3, Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy, Strikingly, osmads3-4 dl-sup6 displayed a severe loss of floral meristem determinacy and produced supernumerary whorls of lodicule-like organs at the forth whorl, suggesting that OsMADS3 and DL synergistically terminate the floral meristem
- DL, OSMADS3, Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy, Collectively, we propose a model to illustrate the role of OsMADS3, DL, and OsMADS13 in the specification of flower organ identity and meristem determinacy in rice
- OsMADS13, OSMADS3, Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy, Here, we report the interactions of rice (Oryza sativa) floral homeotic genes, OsMADS3 (a C-class gene), OsMADS13 (a D-class gene), and DROOPING LEAF (DL), in specifying floral organ identities and floral meristem determinacy
- OsMADS13, OSMADS3, Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy, osmads13-3 osmads3-4 displayed a loss of floral meristem determinacy and generated abundant carpelloid structures containing severe defective ovules in the flower center, which were not detectable in the single mutant
- OsMADS13, OSMADS3, Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy, In addition, in situ hybridization and yeast two-hybrid analyses revealed that OsMADS13 and OsMADS3 did not regulate each other’s transcription or interact at the protein level
- OsMADS13, OSMADS3, Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy, This indicates that OsMADS3 plays a synergistic role with OsMADS13 in both ovule development and floral meristem termination
- OsMADS13, OSMADS3, Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy, Collectively, we propose a model to illustrate the role of OsMADS3, DL, and OsMADS13 in the specification of flower organ identity and meristem determinacy in rice
- OsMADS13, OSMADS3, Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy, Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy
- OSMADS3, OSMADS58, Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa, To examine the conservation and diversification of C-class gene function in monocots, we analyzed two C-class genes in rice (Oryza sativa), OSMADS3 and OSMADS58, which may have arisen by gene duplication before divergence of rice and maize (Zea mays)
- OSMADS3, OSMADS58, Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa, , the functions regulated by AG have been partially partitioned into two paralogous genes, OSMADS3 and OSMADS58, which were produced by a recent gene duplication event in plant evolution)
- OSMADS3, OSMADS58, Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa, Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa
- OsMADS1~LHS1~AFO, OSMADS3, Interactions of OsMADS1 with Floral Homeotic Genes in Rice Flower Development., The physical and genetic interaction of OsMADS1 and OsMADS3 is essential for floral meristem activity maintenance and organ identity specification; while OsMADS1 physically and genetically interacts with OsMADS58 in regulating floral meristem determinacy and suppressing spikelet meristem reversion
- MEL2, OSMADS3, Rice MEL2, the RNA recognition motif RRM protein, binds in vitro to meiosis-expressed genes containing U-rich RNA consensus sequences in the 3’-UTR., Of 249 genes that conserved the consensus in their 3’-UTR, 13 genes spatiotemporally co-expressed with MEL2 in meiotic flowers, and included several genes whose function was supposed in meiosis; such as Replication protein A and OsMADS3
- FON2~FON4~TG1, OSMADS3, Genetic Enhancer Analysis Reveals that FLORAL ORGAN NUMBER2 and OsMADS3 Cooperatively Regulate Maintenance and Determinacy of the Flower Meristem in Rice., Disruption of OsMADS3 in the fon2 mutant by CRISPR-Cas9 technology caused a flower phenotype similar to that of 2B-424, confirming that the gene responsible for enhancement of fon2 was OsMADS3
- FON2~FON4~TG1, OSMADS3, Genetic Enhancer Analysis Reveals that FLORAL ORGAN NUMBER2 and OsMADS3 Cooperatively Regulate Maintenance and Determinacy of the Flower Meristem in Rice., Morphological analysis showed that the fon2 and osmads3 mutations synergistically affected pistil development and FM determinacy
- NSG, OSMADS3, nonstop glumes nsg, a novel mutant affects spikelet development in rice, The expression of OsMADS4, OsMADS16, DL and OsMADS3 decreased distinctly, and OsMADS1 increased in nsg panicle, suggests that NSG affected spikelet development through influencing the expression of floral hometic genes
Prev Next