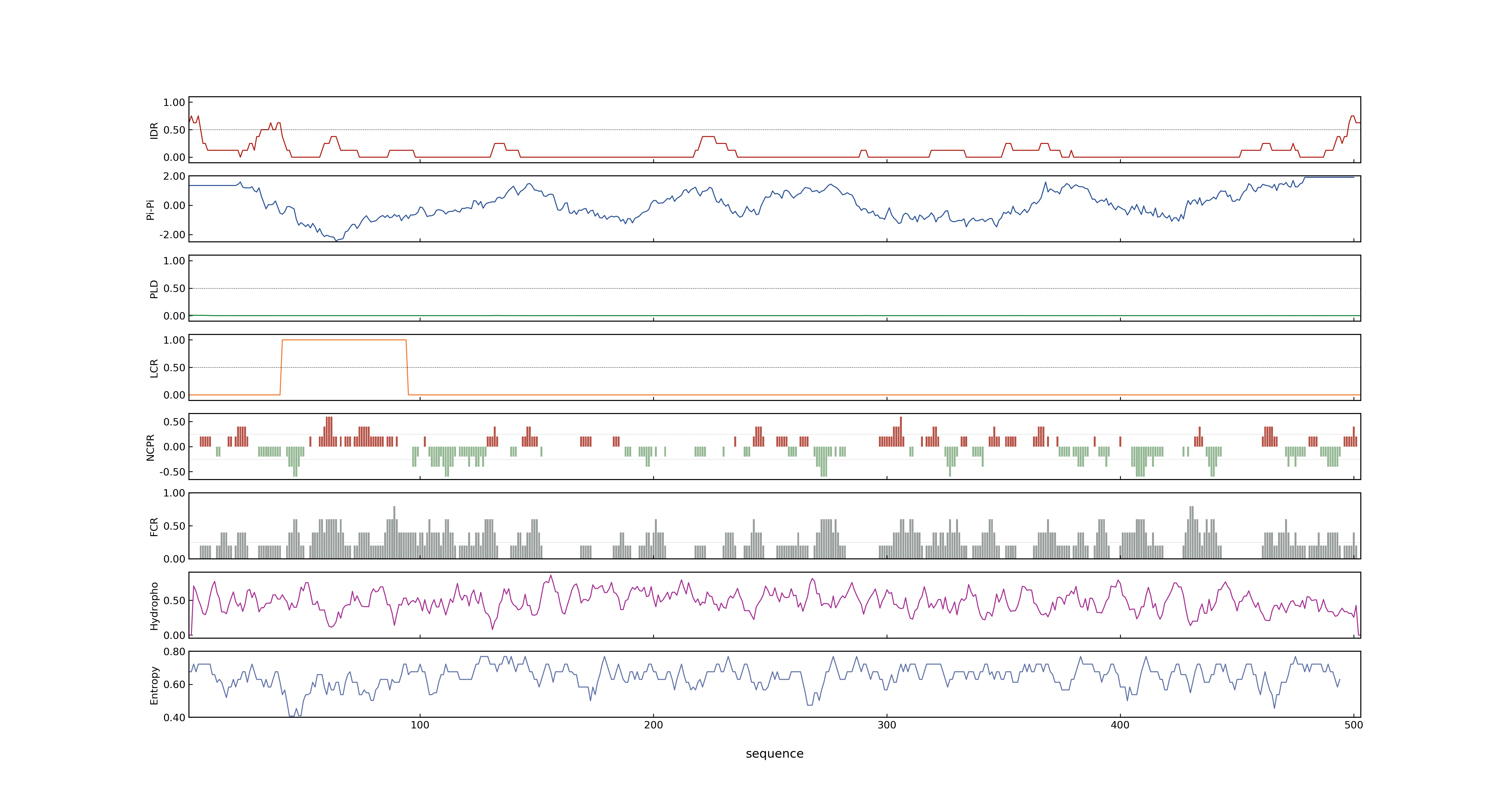
- Information
- Symbol: OsBADH2,fgr
- MSU: LOC_Os08g32870
- RAPdb: Os08g0424500
- PSP score
- LOC_Os08g32870.1: 0.0034
- PLAAC score
- LOC_Os08g32870.1: 0
- pLDDT score
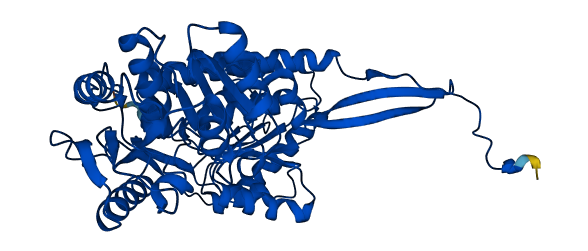
- 97.83
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os08g32870.1: 0.00241206
- MolPhase Result
- Publication
- Aroma in rice: genetic analysis of a quantitative trait, 1996, Theor Appl Genet.
- OsBADH1 is possibly involved in acetaldehyde oxidation in rice plant peroxisomes, 2009, FEBS Lett.
- Dissecting substrate specificity of two rice BADH isoforms: Enzyme kinetics, docking and molecular dynamics simulation studies, 2012, Biochimie.
- Haplotype variation at Badh2, the gene determining fragrance in rice, 2012, Genomics.
- RFLP tagging of a gene for aroma in rice, 1992, Theor Appl Genet.
- Characterization of the major fragance gene from an aromatic japonica rice and analysis of its diversity in Asian cultivated rice, 2008, Theor Appl Genet.
- RNAi-directed downregulation of OsBADH2 results in aroma 2-acetyl-1-pyrroline production in rice Oryza sativa L., 2008, BMC Plant Biol.
- Purification, crystallization and preliminary X-ray analysis of recombinant betaine aldehyde dehydrogenase 2 OsBADH2, a protein involved in jasmine aroma, from Thai fragrant rice Oryza sativa L., 2011, Acta Crystallogr Sect F Struct Biol Cryst Commun.
- The gene for fragrance in rice, 2005, Plant Biotechnol J.
- Badh2, encoding betaine aldehyde dehydrogenase, inhibits the biosynthesis of 2-acetyl-1-pyrroline, a major component in rice fragrance, 2008, Plant Cell.
- The origin and evolution of fragrance in rice Oryza sativa L., 2009, Proc Natl Acad Sci U S A.
- Biochemical and enzymatic study of rice BADH wild-type and mutants: an insight into fragrance in rice, 2011, Protein J.
- Inactivation of an aminoaldehyde dehydrogenase is responsible for fragrance in rice, 2008, Plant Mol Biol.
- Creation of fragrant rice by targeted knockout of the OsBADH2 gene using TALEN technology., 2015, Plant Biotechnol J.
- Resequencing Reveals Different Domestication Rate for BADH1 and BADH2 in Rice Oryza sativa., 2015, PLoS One.
- Genbank accession number
- Key message
- RESULTS: The results showed that multiple mutations identical to fgr allele occur in the 13 fragrant rice accessions across China; OsBADH2 is expressed constitutively, with less expression abundance in mature roots; the disrupted OsBADH2 by RNA interference leads to significantly increased 2-acetyl-1-pyrroline production
- Connection
- BAD1~OsBADH1, OsBADH2~fgr, OsBADH1 is possibly involved in acetaldehyde oxidation in rice plant peroxisomes, We found that OsBADH1 catalyzes the oxidation of acetaldehyde efficiently, while the activity of OsBADH2 is extremely low
- BAD1~OsBADH1, OsBADH2~fgr, Dissecting substrate specificity of two rice BADH isoforms: Enzyme kinetics, docking and molecular dynamics simulation studies, Fragrance rice (Oryza sativa) contains two isoforms of BADH, named OsBADH1 and OsBADH2
- BAD1~OsBADH1, OsBADH2~fgr, Dissecting substrate specificity of two rice BADH isoforms: Enzyme kinetics, docking and molecular dynamics simulation studies, OsBADH1 is implicated in acetaldehyde oxidation in rice plant peroxisomes, while the non-functional OsBADH2 is believed to be involved in the accumulation of 2-acetyl-1-pyrroline, the major compound of aroma in fragrance rice
- BAD1~OsBADH1, OsBADH2~fgr, Dissecting substrate specificity of two rice BADH isoforms: Enzyme kinetics, docking and molecular dynamics simulation studies, Consistent with our previous study, kinetics data indicated that the enzymes catalyze the oxidation of GAB-ald more efficiently than Bet-ald and the OsBADH1 W172F and OsBADH2 W170F mutants displayed a higher catalytic efficiency towards GAB-ald
- BAD1~OsBADH1, OsBADH2~fgr, Dissecting substrate specificity of two rice BADH isoforms: Enzyme kinetics, docking and molecular dynamics simulation studies, The amino acid residues, E262, L263, C296 and W461 of OsBADH1 and E260, L261, C294 and W459 of OsBADH2 located within 5 A of the OsBADH active site mainly interacted with GAB-ald forming strong hydrogen bonds in both OsBADH isoforms
- BAD1~OsBADH1, OsBADH2~fgr, Dissecting substrate specificity of two rice BADH isoforms: Enzyme kinetics, docking and molecular dynamics simulation studies, Residues W163, N164, Q294, C296 and F397 of OsBADH1-Bet-ald and Y163, M167, W170, E260, S295 and C453 of OsBADH2-Bet-ald formed the main interaction sites while E260 showed an interaction energy of -14
- BAD1~OsBADH1, OsBADH2~fgr, Dissecting substrate specificity of two rice BADH isoforms: Enzyme kinetics, docking and molecular dynamics simulation studies, Unconserved A290 in OsBADH1 and W288 in OsBADH2 appeared to be important for substrate recognition similar to that observed in PsAMADHs
Prev Next