- Information
- Symbol: OsEREBP2,ERF99
- MSU: LOC_Os01g64790
- RAPdb: Os01g0868000
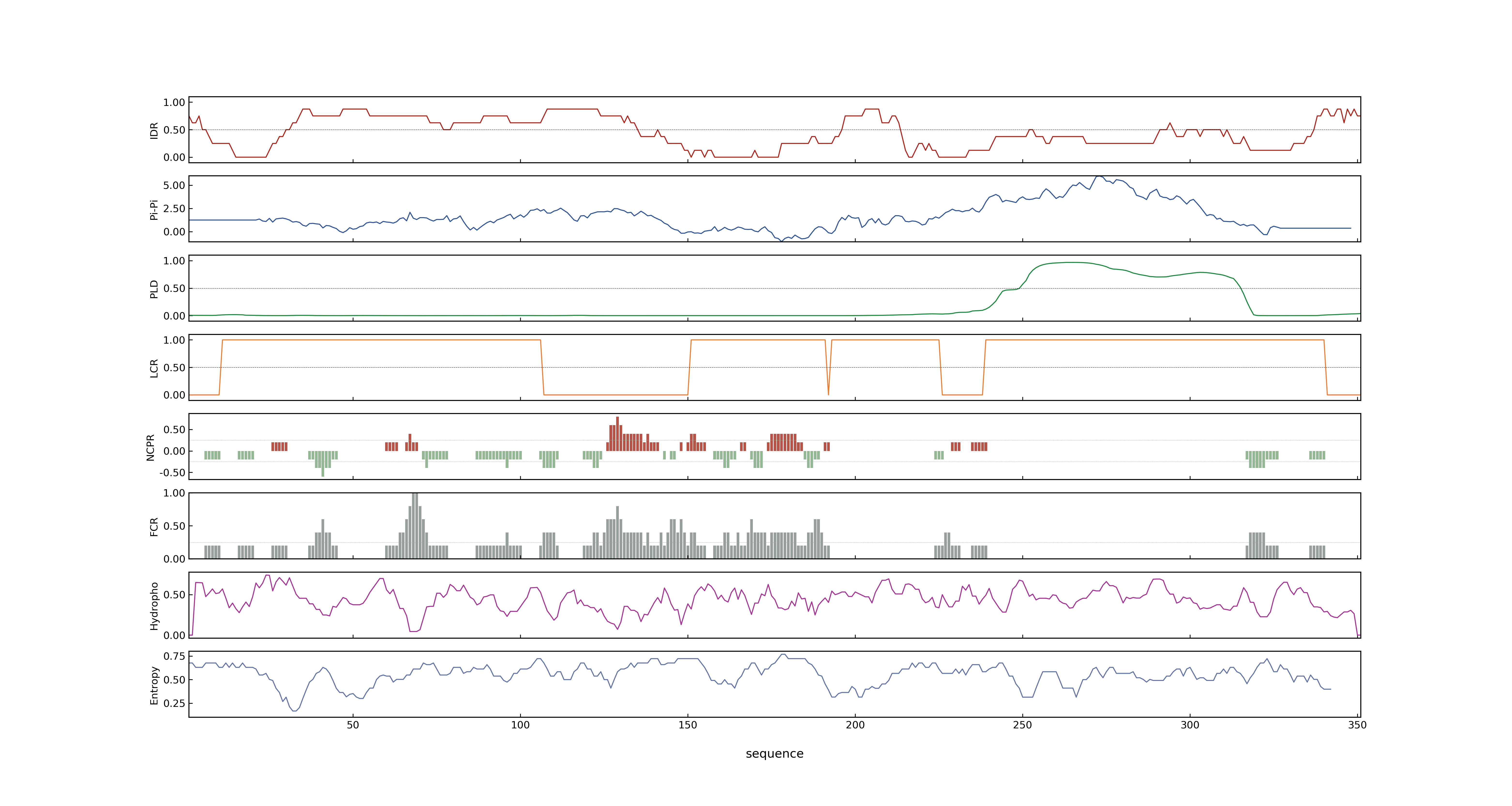
- PSP score
- LOC_Os01g64790.1: 0.6324
- PLAAC score
- LOC_Os01g64790.1: 0
- pLDDT score
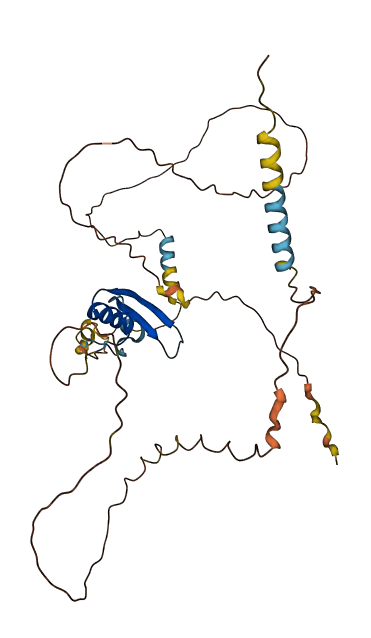
- 54.32
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os01g64790.1: 0.99777871
- MolPhase Result
- Publication
-
Genbank accession number
- Key message
- On the other hand, the OsEREBP2 transcript level increased after cold, ABA, drought and high salinity treatments, indicating that OsEREBP2 may play a central role mediating the response to different abiotic stresses
- Gene expression analysis in rice varieties with contrasting salt tolerance further suggests that OsEREBP2 is involved in salt stress response in rice
- Thereby, two transcription factors (TFs), OsEREBP1 and OsEREBP2, belonging to the AP2/ERF family were identified
- Connection
- OsEREBP1, OsEREBP2~ERF99, OsRMC, a negative regulator of salt stress response in rice, is regulated by two AP2/ERF transcription factors, Thereby, two transcription factors (TFs), OsEREBP1 and OsEREBP2, belonging to the AP2/ERF family were identified
- OsEREBP2~ERF99, OsHOS1, The rice E3 ubiquitin ligase OsHOS1 modulates the expression of OsRMC, a gene involved in root mechano-sensing, through the interaction with two ERF transcription factors., Using the yeast two-hybrid system and BiFC assays we showed that OsHOS1 interacts with two ERF transcription factors (TFs), OsEREBP1 and OsEREBP2, known to regulate OsRMC gene expression
- OsEREBP2~ERF99, OsHOS1, The rice E3 ubiquitin ligase OsHOS1 modulates the expression of OsRMC, a gene involved in root mechano-sensing, through the interaction with two ERF transcription factors., In addition, we showed that OsHOS1 affects the stability of both TFs in a proteasome dependent way, suggesting that this E3-ubiquitin ligase targets OsEREBP1 and OsEREBP2 for degradation
Prev Next