- Information
- Symbol: OsGAPDH
- MSU: LOC_Os08g34210
- RAPdb: Os08g0440800
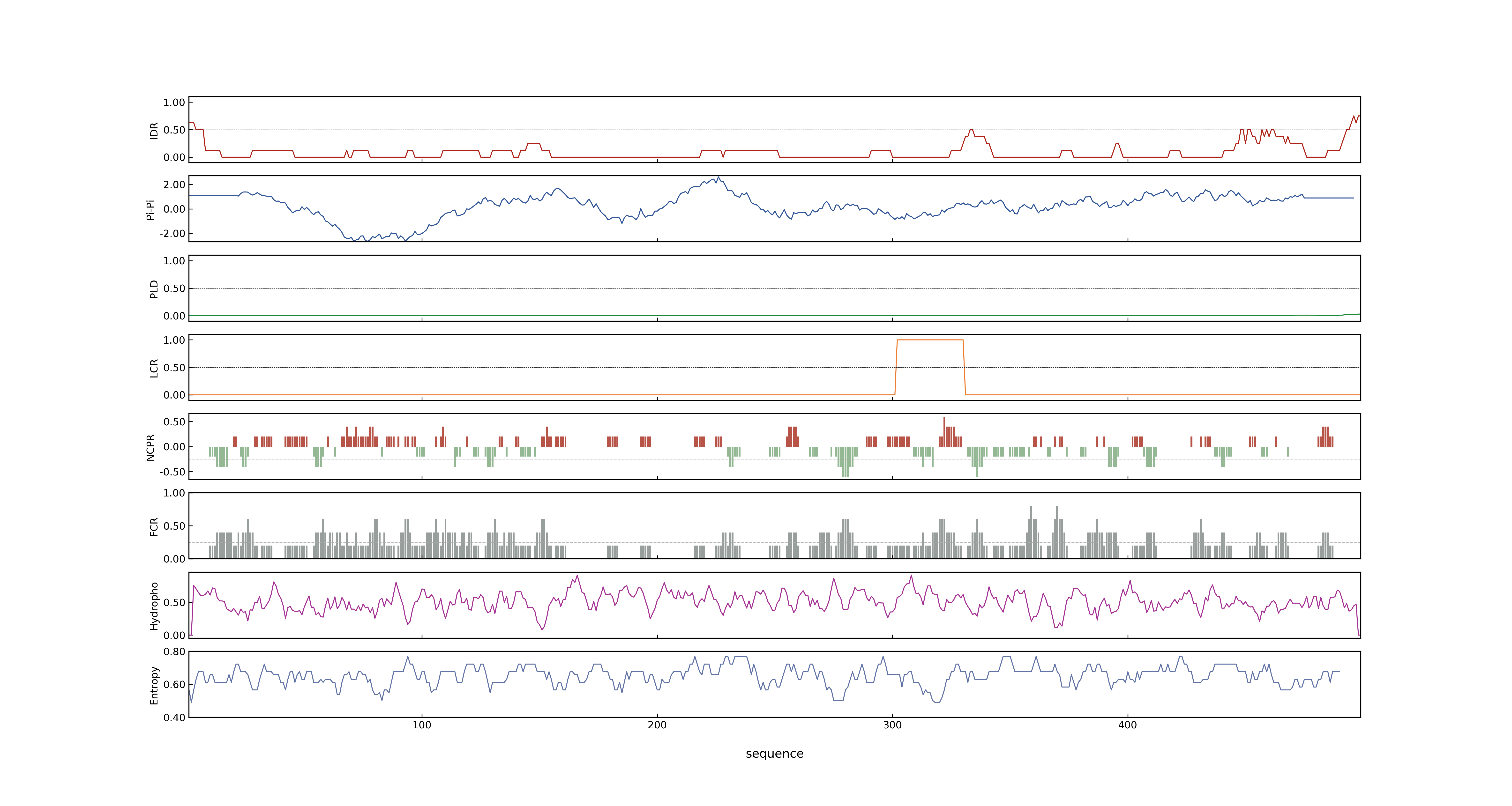
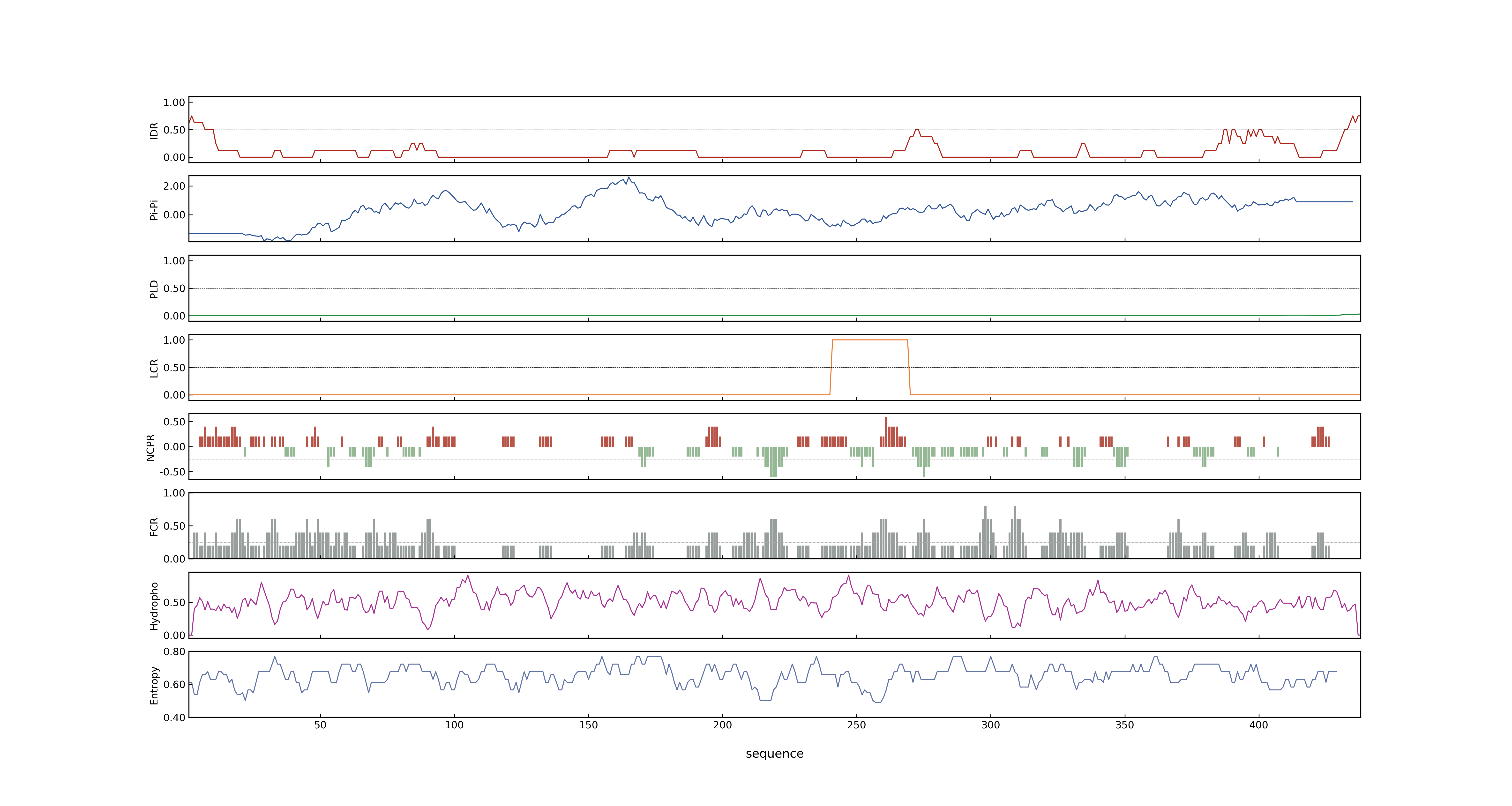
- PSP score
- LOC_Os08g34210.2: 0.0289
- LOC_Os08g34210.1: 0.0204
- PLAAC score
- LOC_Os08g34210.2: 0
- LOC_Os08g34210.1: 0
- pLDDT score
- 97.14
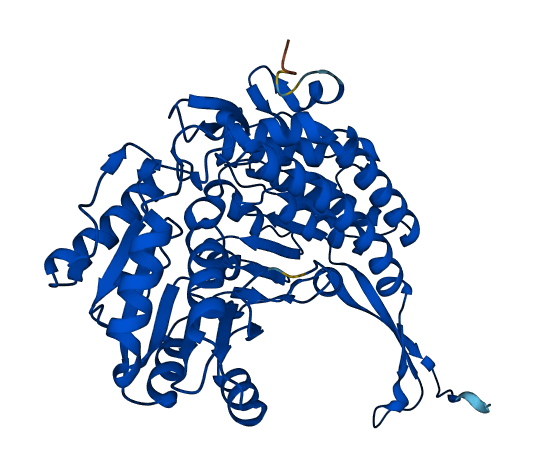
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os08g34210.1: 0.03259879
- LOC_Os08g34210.2: 0.00837867
- MolPhase Result
- Publication
- Crystal structures of rice Oryza sativa glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and sulfate suggest involvement of Phe37 in NAD binding for catalysis, 2012, Plant Mol Biol.
- Molecular cloning, characterization, expression and chromosomal location of OsGAPDH, a submergence responsive gene in rice Oryza sativa L., 2002, Theor Appl Genet.
-
Genbank accession number
- Key message
- Plants exposed to drought, submergence and ABA treatment showed an increased accumulation of OsGAPDH transcripts
- Tissue-specific expression of OsGAPDH indicated that the high level of mRNA was detected in the panicle
- Cytosolic Oryza sativa glyceraldehyde-3-phosphate dehydrogenase (OsGAPDH), the enzyme involved in the ubiquitous glycolysis, catalyzes the oxidative phosphorylation of glyceraldehyde-3-phosphate to 1,3-biphosphoglycerate (BPG) using nicotinamide adenine dinucleotide (NAD) as an electron acceptor
- NAD binds to each OsGAPDH subunits with some residues forming positively charged grooves that attract sulfate anions, as a simulation of phosphate groups in the product BPG
- One of the clones, OsGAPDH, represented a gene that was expressed at high level during 12-h submergence
- Molecular cloning, characterization, expression and chromosomal location of OsGAPDH, a submergence responsive gene in rice ( Oryza sativa L.)
- Connection
Prev Next