- Information
- Symbol: OsGF14e,GF14e,GID2
- MSU: LOC_Os02g36974
- RAPdb: Os02g0580300
- PSP score
- LOC_Os02g36974.1: 0.0325
- LOC_Os02g36974.6: 0.0764
- LOC_Os02g36974.2: 0.0325
- LOC_Os02g36974.5: 0.0488
- LOC_Os02g36974.3: 0.0325
- LOC_Os02g36974.4: 0.0325
- PLAAC score
- LOC_Os02g36974.1: 0
- LOC_Os02g36974.6: 0
- LOC_Os02g36974.2: 0
- LOC_Os02g36974.5: 0
- LOC_Os02g36974.3: 0
- LOC_Os02g36974.4: 0
- pLDDT score
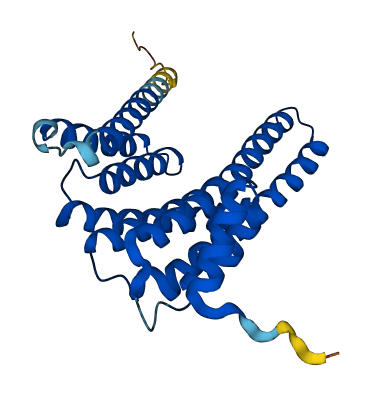
- 92.86
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os02g36974.1: 0.01805595
- LOC_Os02g36974.2: 0.01805595
- LOC_Os02g36974.3: 0.01805595
- LOC_Os02g36974.4: 0.01805595
- LOC_Os02g36974.5: 0.00049457
- LOC_Os02g36974.6: 0.00073106
- MolPhase Result
- Publication
- Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice, 2010, Plant Cell.
- The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress, 2006, DNA Res.
- Gibberellin homeostasis and plant height control by EUI and a role for gibberellin in root gravity responses in rice, 2008, Cell Res.
- GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice, 2004, The Plant Journal.
- Rice 14-3-3 protein GF14e negatively affects cell death and disease resistance, 2011, Plant J.
- Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant, 2008, Plant Cell.
- Accumulation of Phosphorylated Repressor for Gibberellin Signaling in an F-box Mutant, 2003, Science.
- OsGF14e positively regulates panicle blast resistance in rice., 2016, Biochem Biophys Res Commun.
- Rice black streaked dwarf virus P7-2 forms a SCF complex through binding to Oryza sativa SKP1-like proteins, and interacts with GID2 involved in the gibberellin pathway., 2017, PLoS One.
- Genbank accession number
- Key message
- Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant
- In gid2, a repressor for GA signaling, SLR1, was highly accumulated in a phosphorylated form and GA increased its concentration, whereas SLR1 was rapidly degraded by GA through ubiquitination in the wild type
- We conclude that GID2 is a positive regulator of GA signaling and that regulated degradation of SLR1 is initiated through GA-dependent phosphorylation and finalized by an SCF(GID2)-proteasome pathway
- Enhanced resistance was correlated with GF14e silencing prior to and after development of the LM phenotype, higher basal expression of a defense response peroxidase gene (POX22
- Together, our findings suggest that GF14e negatively affects the induction of plant defense response genes, cell death and broad-spectrum resistance in rice
- Disturbing GA homeostasis affected the expression of the GA signaling genes GID1 (GIBBERELLIN INSENSITIVE DWARF 1), GID2 and SLR1
- We isolated and characterized a rice GA-insensitive dwarf mutant, gid2
- It is assumed that interaction between GIBBERELLIN INSENSITIVE DWARF1 (GID1) and the N-terminal DELLA/TVHYNP motif of SLR1 triggers F-box protein GID2-mediated SLR1 degradation
- To determine whether the GF14e gene plays a direct role in resistance to disease in rice, we suppressed its expression by RNAi silencing
- Rice 14-3-3 protein (GF14e) negatively affects cell death and disease resistance
- GF14e-silenced rice plants showed high levels of resistance to a virulent strain of Xoo compared with plants that were not silenced
- GA perception by GID1 causes SLR1 protein degradation involving the F-box protein GID2; this triggers GA-associated responses such as shoot elongation and seed germination
- Previously, we identified a rice F-box protein, gibberellin-insensitive dwarf2 (GID2), which is essential for GA-mediated DELLA protein degradation
- GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice
- The level of SLR1 protein in gid2 was decreased by loss of GID1 function or treatment with a GA biosynthesis inhibitor, and dwarfism was enhanced
- These results indicate that derepression of SLR1 repressive activity can be accomplished by GA and GID1 alone and does not require F-box (GID2) function
- Evidence for GA signaling without GID2 was also provided by the expression behavior of GA-regulated genes such as GA-20oxidase1, GID1, and SLR1 in the gid2 mutant
- An exception is the GA-insensitive F-box mutant gid2, which shows milder dwarfism than mutants such as gid1 and cps even though it accumulates higher levels of SLR1
- Conversely, overproduction of GID1 or treatment with GA(3) increased the SLR1 level in gid2 and reduced dwarfism
- OsGF14e positively regulates panicle blast resistance in rice.
- Rice black streaked dwarf virus P7-2 forms a SCF complex through binding to Oryza sativa SKP1-like proteins, and interacts with GID2 involved in the gibberellin pathway.
- Overall, these results indicated that P7-2 functioned as a component of the SCF complex in rice, and interaction of P7-2 with GID2 implied possible roles of the GA signaling pathway during RBSDV infection
- Connection
- OsGF14e~GF14e~GID2, SLR1~OsGAI, Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice, It is assumed that interaction between GIBBERELLIN INSENSITIVE DWARF1 (GID1) and the N-terminal DELLA/TVHYNP motif of SLR1 triggers F-box protein GID2-mediated SLR1 degradation
- OsGF14e~GF14e~GID2, SLR1~OsGAI, Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice, The GA-dependent degradation of SLR1(G576V) was reduced in Slr1-d4, and compared with SLR1, SLR1(G576V) showed reduced interaction with GID1 and almost none with GID2 when tested in yeast cells
- OsGF14e~GF14e~GID2, SLR1~OsGAI, Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice, These results suggest that the stable interaction of GID1-SLR1 through the GRAS domain is essential for the recognition of SLR1 by GID2
- OsGF14e~GF14e~GID2, SLR1~OsGAI, Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice, We propose that when the DELLA/TVHYNP motif of SLR1 binds with GID1, it enables the GRAS domain of SLR1 to interact with GID1 and that the stable GID1-SLR1 complex is efficiently recognized by GID2
- GID1~OsGID1, OsGF14e~GF14e~GID2, Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice, It is assumed that interaction between GIBBERELLIN INSENSITIVE DWARF1 (GID1) and the N-terminal DELLA/TVHYNP motif of SLR1 triggers F-box protein GID2-mediated SLR1 degradation
- GID1~OsGID1, OsGF14e~GF14e~GID2, Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice, The GA-dependent degradation of SLR1(G576V) was reduced in Slr1-d4, and compared with SLR1, SLR1(G576V) showed reduced interaction with GID1 and almost none with GID2 when tested in yeast cells
- GID1~OsGID1, OsGF14e~GF14e~GID2, Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice, These results suggest that the stable interaction of GID1-SLR1 through the GRAS domain is essential for the recognition of SLR1 by GID2
- GID1~OsGID1, OsGF14e~GF14e~GID2, Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice, We propose that when the DELLA/TVHYNP motif of SLR1 binds with GID1, it enables the GRAS domain of SLR1 to interact with GID1 and that the stable GID1-SLR1 complex is efficiently recognized by GID2
- GF14f, OsGF14e~GF14e~GID2, The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress, At least four rice GF14 genes, GF14b, GF14c, GF14e and Gf14f were differentially regulated in the interactions of rice-Magnaporthe grisea and rice-Xanthomonas oryzae pv
- GF14b~OsGF14b, OsGF14e~GF14e~GID2, The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress, At least four rice GF14 genes, GF14b, GF14c, GF14e and Gf14f were differentially regulated in the interactions of rice-Magnaporthe grisea and rice-Xanthomonas oryzae pv
- GF14c, OsGF14e~GF14e~GID2, The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress, At least four rice GF14 genes, GF14b, GF14c, GF14e and Gf14f were differentially regulated in the interactions of rice-Magnaporthe grisea and rice-Xanthomonas oryzae pv
- OsGF14e~GF14e~GID2, SLR1~OsGAI, Gibberellin homeostasis and plant height control by EUI and a role for gibberellin in root gravity responses in rice, Disturbing GA homeostasis affected the expression of the GA signaling genes GID1 (GIBBERELLIN INSENSITIVE DWARF 1), GID2 and SLR1
- GID1~OsGID1, OsGF14e~GF14e~GID2, Gibberellin homeostasis and plant height control by EUI and a role for gibberellin in root gravity responses in rice, Disturbing GA homeostasis affected the expression of the GA signaling genes GID1 (GIBBERELLIN INSENSITIVE DWARF 1), GID2 and SLR1
- OsGF14e~GF14e~GID2, SLR1~OsGAI, GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice, Furthermore, an in vitro pull-down assay revealed that GID2 specifically interacted with the phosphorylated Slender Rice 1 (SLR1)
- OsGF14e~GF14e~GID2, SLR1~OsGAI, GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice, Taken these results together, we conclude that the phosphorylated SLR1 is caught by the SCFGID2 complex through an interacting affinity between GID2 and phosphorylated SLR1, triggering the ubiquitin-mediated degradation of SLR1
- OsGF14e~GF14e~GID2, SLR1~OsGAI, GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice, GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice
- GID1~OsGID1, OsGF14e~GF14e~GID2, Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant, GA perception by GID1 causes SLR1 protein degradation involving the F-box protein GID2; this triggers GA-associated responses such as shoot elongation and seed germination
- GID1~OsGID1, OsGF14e~GF14e~GID2, Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant, An exception is the GA-insensitive F-box mutant gid2, which shows milder dwarfism than mutants such as gid1 and cps even though it accumulates higher levels of SLR1
- GID1~OsGID1, OsGF14e~GF14e~GID2, Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant, The level of SLR1 protein in gid2 was decreased by loss of GID1 function or treatment with a GA biosynthesis inhibitor, and dwarfism was enhanced
- GID1~OsGID1, OsGF14e~GF14e~GID2, Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant, Conversely, overproduction of GID1 or treatment with GA(3) increased the SLR1 level in gid2 and reduced dwarfism
- GID1~OsGID1, OsGF14e~GF14e~GID2, Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant, These results indicate that derepression of SLR1 repressive activity can be accomplished by GA and GID1 alone and does not require F-box (GID2) function
- GID1~OsGID1, OsGF14e~GF14e~GID2, Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant, Evidence for GA signaling without GID2 was also provided by the expression behavior of GA-regulated genes such as GA-20oxidase1, GID1, and SLR1 in the gid2 mutant
- OsGF14e~GF14e~GID2, SLR1~OsGAI, Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant, GA perception by GID1 causes SLR1 protein degradation involving the F-box protein GID2; this triggers GA-associated responses such as shoot elongation and seed germination
- OsGF14e~GF14e~GID2, SLR1~OsGAI, Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant, An exception is the GA-insensitive F-box mutant gid2, which shows milder dwarfism than mutants such as gid1 and cps even though it accumulates higher levels of SLR1
- OsGF14e~GF14e~GID2, SLR1~OsGAI, Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant, The level of SLR1 protein in gid2 was decreased by loss of GID1 function or treatment with a GA biosynthesis inhibitor, and dwarfism was enhanced
- OsGF14e~GF14e~GID2, SLR1~OsGAI, Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant, Conversely, overproduction of GID1 or treatment with GA(3) increased the SLR1 level in gid2 and reduced dwarfism
- OsGF14e~GF14e~GID2, SLR1~OsGAI, Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant, These results indicate that derepression of SLR1 repressive activity can be accomplished by GA and GID1 alone and does not require F-box (GID2) function
- OsGF14e~GF14e~GID2, SLR1~OsGAI, Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant, Evidence for GA signaling without GID2 was also provided by the expression behavior of GA-regulated genes such as GA-20oxidase1, GID1, and SLR1 in the gid2 mutant
- OsGF14e~GF14e~GID2, SLR1~OsGAI, Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant, Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant
- OsCPK21, OsGF14e~GF14e~GID2, Calcium-dependent protein kinase 21 phosphorylates 14-3-3 proteins in response to ABA signaling and salt stress in rice., In the present study, we performed yeast two-hybrid screening, glutathione S-transferase pull-down, co-immunoprecipitation, and bimolecular fluorescence complementation assays to confirm the interaction between OsCPK21 and one of its putative targets, Os14-3-3 (OsGF14e)
- OsCPK21, OsGF14e~GF14e~GID2, Calcium-dependent protein kinase 21 phosphorylates 14-3-3 proteins in response to ABA signaling and salt stress in rice., We used an in<U+00A0>vitro kinase assay and site-directed mutagenesis to verify that OsCPK21 phosphorylates OsGF14e at Tyr-138
- OsCPK21, OsGF14e~GF14e~GID2, Calcium-dependent protein kinase 21 phosphorylates 14-3-3 proteins in response to ABA signaling and salt stress in rice., These results suggest that OsCPK21 phosphorylates OsGF14e to facilitate the response to ABA and salt stress
Prev Next