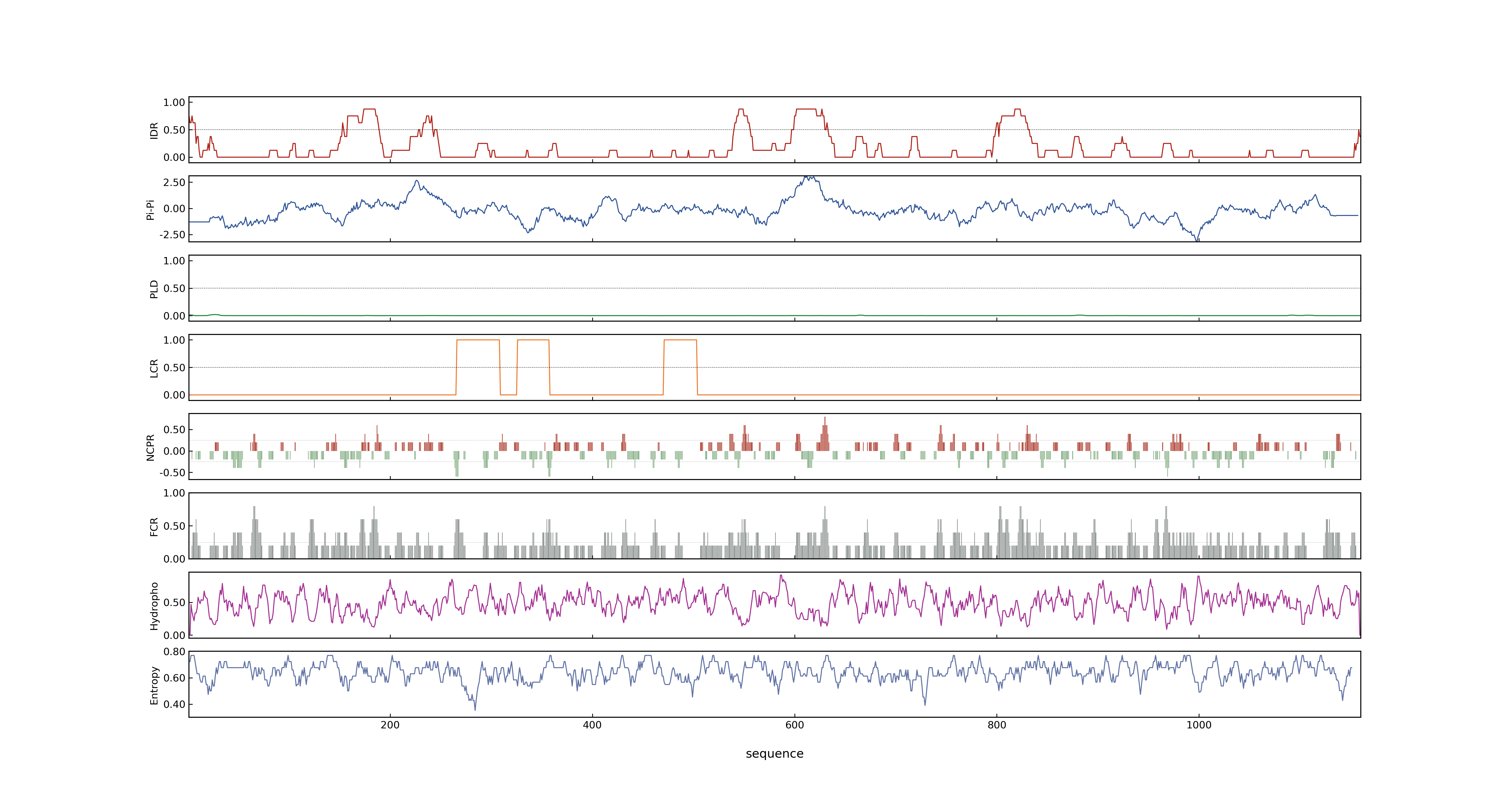
- Information
- Symbol: OsGI
- MSU: LOC_Os01g08700
- RAPdb: Os01g0182600
- PSP score
- LOC_Os01g08700.1: 0.3948
- LOC_Os01g08700.4: 0.3948
- LOC_Os01g08700.3: 0.3948
- LOC_Os01g08700.8: 0.7239
- LOC_Os01g08700.2: 0.3948
- LOC_Os01g08700.5: 0.3948
- LOC_Os01g08700.6: 0.3948
- PLAAC score
- LOC_Os01g08700.1: 0
- LOC_Os01g08700.4: 0
- LOC_Os01g08700.3: 0
- LOC_Os01g08700.8: 0
- LOC_Os01g08700.2: 0
- LOC_Os01g08700.5: 0
- LOC_Os01g08700.6: 0
(Zheng et al., New Phytol, 2019, 224:306-320)
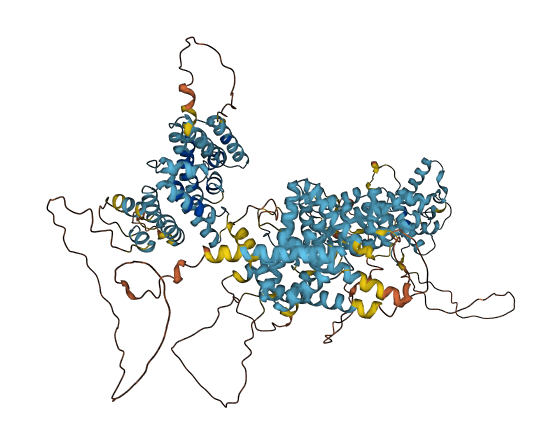
- pLDDT score
- 66.85
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os01g08700.1: 0.97666275
- LOC_Os01g08700.2: 0.97666275
- LOC_Os01g08700.3: 0.97666275
- LOC_Os01g08700.4: 0.97666275
- LOC_Os01g08700.5: 0.97666275
- LOC_Os01g08700.6: 0.97666275
- LOC_Os01g08700.8: 0.98647618
- MolPhase Result
- Publication
- An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, 2011, N Biotechnol.
- Functional characterization of rice OsDof12, 2009, Planta.
- Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field, 2011, Plant Cell.
- Circadian regulation of rice Oryza sativa L. CONSTANS-like gene transcripts, 2004, Mol Cells.
- Adaptation of photoperiodic control pathways produces short-day flowering in rice, 2003, Nature.
- OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, 2009, Plant Cell Environ.
- Physiological significance of the plant circadian clock in natural field conditions, 2012, Plant Cell Environ.
- Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 SOC1/AGL20 ortholog in rice, 2004, Plant J.
- OsELF3 is involved in circadian clock regulation for promoting flowering under long-day conditions in rice, 2013, Mol Plant.
- A pair of floral regulators sets critical day length for Hd3a florigen expression in rice, 2010, Nat Genet.
- Isolation of Rice Genes Possibly Involved in the Photoperiodic Control of Flowering by a Fluorescent Differential Display Method, 2002, Plant and Cell Physiology.
- Identification of dynamin as an interactor of rice GIGANTEA by tandem affinity purification TAP, 2008, Plant Cell Physiol.
- OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a, 2007, Plant Physiol.
- Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date3, 2014, Plant Physiol.
- A study of phytohormone biosynthetic gene expression using a circadian clock-related mutant in rice, 2011, Plant Signal Behav.
- Mutation of OsGIGANTEA Leads to Enhanced Tolerance to Polyethylene Glycol-Generated Osmotic Stress in Rice., 2016, Front Plant Sci.
- Genbank accession number
- Key message
- Time-course metabolome analyses of leaves revealed no trends of change in primary metabolites in osgi plants, and net photosynthetic rates and grain yields were not affected
- Consistently, osgi-1 plants showed semi-dwarf phenotype with reduced internode and leaf sheath elongation
- The S3574 and C60910 genes were expressed to similar extents during the vegetative and reproductive phases, like OsGI
- Here we found that mRNA levels of a few major genes encoding GA2-oxidase which can inactivate bioactive gibberellins (GAs) were remarkably increased in osgi-1 plants
- Overexpression of OsGI, an orthologue of the Arabidopsis GIGANTEA (GI) gene in transgenic rice, caused late flowering under both SD and LD conditions
- Hd3a expression is induced by Ehd1 (Early heading date 1) expression when blue light coincides with the morning phase set by OsGIGANTEA(OsGI)-dependent circadian clocks
- In rice, OsGI, Hd1 and Hd3a were identified as orthologs of GI, CO and FT, respectively, and are also important regulators of flowering
- Further investigations showed that the expression of OsGI and Ghd7 was up-regulated in the oself3 mutant, indicating that OsELF3 acts as a negative regulator upstream of OsGI and Ghd7 in the flowering-time control under long-day conditions
- Transcript levels of OsGI, phytochrome genes, and Early heading date3 (Ehd3), which function upstream of Ghd7, were unchanged in the mutant
- Recently, an extensive time-course transcriptome analysis of rice (Oryza sativa) leaves in natural field conditions revealed that OsGIGANTEA, the sole rice ortholog of the Arabidopsis GIGANTEA gene, governs the robust diurnal rhythm of rice leaf transcriptomes even under natural diurnal conditions; rice Osgi mutants exhibited severely defective transcriptome rhythms under strong diurnal changes in environmental cues
- Flowering genes downstream of OsPRR1 such as OsGI and Hd1 were down regulated in the A654 mutant
- Together, these results suggest that overexpression of OsLF might repress expression of OsGI and Hd1 by competing with OsPRR1 in interacting with OsPIL13 and OsPIL15 and thus induce late flowering
- Transcript levels of three flowering regulators-Ehd1, OsMADS14, and Hd3a-were decreased in these mutants, whereas those of OsGI and Hd1 were unchanged
- To further investigate the relationship with other flowering promoters, we generated transgenic plants in which expression of Ehd1 or OsGI was suppressed
- In summary, OsMADS51 is a novel flowering promoter that transmits a SD promotion signal from OsGI to Ehd1
- The expression patterns of the rice homolog of GI, OsGI, were similar to those of the Arabidopsis GI, suggesting the conservation of some mechanisms for the photoperiodic regulation of flowering between these two species
- Results showed that mutation of OsGI conferred tolerance to osmotic stress generated by polyethylene glycol (PEG), increased proline and sucrose contents, and accelerated stomata movement
- In addition, qRT-PCR and microarray analysis revealed that the transcript abundance of some osmotic stress response genes, such as OsDREB1E, OsAP37, OsAP59, OsLIP9, OsLEA3, OsRAB16A, and OsSalT, was significantly higher in osgi than in WT plants, suggesting that OsGI might be a negative regulator in the osmotic stress response in rice
- Connection
- Hd1, OsGI, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Flowering genes downstream of OsPRR1 such as OsGI and Hd1 were down regulated in the A654 mutant
- Hd1, OsGI, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Together, these results suggest that overexpression of OsLF might repress expression of OsGI and Hd1 by competing with OsPRR1 in interacting with OsPIL13 and OsPIL15 and thus induce late flowering
- OsGI, OsPIL13~OsPIL1, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Together, these results suggest that overexpression of OsLF might repress expression of OsGI and Hd1 by competing with OsPRR1 in interacting with OsPIL13 and OsPIL15 and thus induce late flowering
- OsGI, OsPRR1, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Flowering genes downstream of OsPRR1 such as OsGI and Hd1 were down regulated in the A654 mutant
- OsGI, OsPRR1, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Together, these results suggest that overexpression of OsLF might repress expression of OsGI and Hd1 by competing with OsPRR1 in interacting with OsPIL13 and OsPIL15 and thus induce late flowering
- OsGI, OsLF, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Together, these results suggest that overexpression of OsLF might repress expression of OsGI and Hd1 by competing with OsPRR1 in interacting with OsPIL13 and OsPIL15 and thus induce late flowering
- OsGI, OsPIL15, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Together, these results suggest that overexpression of OsLF might repress expression of OsGI and Hd1 by competing with OsPRR1 in interacting with OsPIL13 and OsPIL15 and thus induce late flowering
- OsGI, OsMADS14, Functional characterization of rice OsDof12, In transgenic lines overexpressing OsDof12, the transcription levels of Hd3a and OsMADS14 were up-regulated under LD conditions but not SD conditions, whereas the expression of Hd1, OsMADS51, Ehd1 and OsGI did not change under LD and SD conditions
- Hd1, OsGI, Functional characterization of rice OsDof12, In transgenic lines overexpressing OsDof12, the transcription levels of Hd3a and OsMADS14 were up-regulated under LD conditions but not SD conditions, whereas the expression of Hd1, OsMADS51, Ehd1 and OsGI did not change under LD and SD conditions
- OsDof12~OsCDF1, OsGI, Functional characterization of rice OsDof12, In transgenic lines overexpressing OsDof12, the transcription levels of Hd3a and OsMADS14 were up-regulated under LD conditions but not SD conditions, whereas the expression of Hd1, OsMADS51, Ehd1 and OsGI did not change under LD and SD conditions
- Ehd1, OsGI, Functional characterization of rice OsDof12, In transgenic lines overexpressing OsDof12, the transcription levels of Hd3a and OsMADS14 were up-regulated under LD conditions but not SD conditions, whereas the expression of Hd1, OsMADS51, Ehd1 and OsGI did not change under LD and SD conditions
- Hd3a, OsGI, Functional characterization of rice OsDof12, In transgenic lines overexpressing OsDof12, the transcription levels of Hd3a and OsMADS14 were up-regulated under LD conditions but not SD conditions, whereas the expression of Hd1, OsMADS51, Ehd1 and OsGI did not change under LD and SD conditions
- OsGI, OsMADS51~OsMADS65, Functional characterization of rice OsDof12, In transgenic lines overexpressing OsDof12, the transcription levels of Hd3a and OsMADS14 were up-regulated under LD conditions but not SD conditions, whereas the expression of Hd1, OsMADS51, Ehd1 and OsGI did not change under LD and SD conditions
- Ghd7, OsGI, OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, On the other hand, mRNA levels of OsGI, Hd1, OsId1, OsDof12, Ghd7, Hd6 and SE5 were unchanged
- Hd1, OsGI, OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, On the other hand, mRNA levels of OsGI, Hd1, OsId1, OsDof12, Ghd7, Hd6 and SE5 were unchanged
- OsDof12~OsCDF1, OsGI, OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, On the other hand, mRNA levels of OsGI, Hd1, OsId1, OsDof12, Ghd7, Hd6 and SE5 were unchanged
- Hd6~CK2, OsGI, OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, On the other hand, mRNA levels of OsGI, Hd1, OsId1, OsDof12, Ghd7, Hd6 and SE5 were unchanged
- Ehd1, OsGI, A pair of floral regulators sets critical day length for Hd3a florigen expression in rice, Hd3a expression is induced by Ehd1 (Early heading date 1) expression when blue light coincides with the morning phase set by OsGIGANTEA(OsGI)-dependent circadian clocks
- Hd3a, OsGI, A pair of floral regulators sets critical day length for Hd3a florigen expression in rice, Hd3a expression is induced by Ehd1 (Early heading date 1) expression when blue light coincides with the morning phase set by OsGIGANTEA(OsGI)-dependent circadian clocks
- Hd1, OsGI, Identification of dynamin as an interactor of rice GIGANTEA by tandem affinity purification TAP, In rice, OsGI, Hd1 and Hd3a were identified as orthologs of GI, CO and FT, respectively, and are also important regulators of flowering
- Hd3a, OsGI, Identification of dynamin as an interactor of rice GIGANTEA by tandem affinity purification TAP, In rice, OsGI, Hd1 and Hd3a were identified as orthologs of GI, CO and FT, respectively, and are also important regulators of flowering
- Ehd1, OsGI, OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a, Transcript levels of three flowering regulators-Ehd1, OsMADS14, and Hd3a-were decreased in these mutants, whereas those of OsGI and Hd1 were unchanged
- Ehd1, OsGI, OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a, In ectopic expression lines, transcript levels of Ehd1, OsMADS14, and Hd3a were increased, but those of OsGI and Hd1 remained the same
- Ehd1, OsGI, OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a, To further investigate the relationship with other flowering promoters, we generated transgenic plants in which expression of Ehd1 or OsGI was suppressed
- Ehd1, OsGI, OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a, In summary, OsMADS51 is a novel flowering promoter that transmits a SD promotion signal from OsGI to Ehd1
- Hd3a, OsGI, OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a, Transcript levels of three flowering regulators-Ehd1, OsMADS14, and Hd3a-were decreased in these mutants, whereas those of OsGI and Hd1 were unchanged
- Hd3a, OsGI, OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a, In ectopic expression lines, transcript levels of Ehd1, OsMADS14, and Hd3a were increased, but those of OsGI and Hd1 remained the same
- OsGI, OsMADS14, OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a, Transcript levels of three flowering regulators-Ehd1, OsMADS14, and Hd3a-were decreased in these mutants, whereas those of OsGI and Hd1 were unchanged
- OsGI, OsMADS14, OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a, In ectopic expression lines, transcript levels of Ehd1, OsMADS14, and Hd3a were increased, but those of OsGI and Hd1 remained the same
- OsGI, OsMADS51~OsMADS65, OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a, However, in OsGI antisense plants, the OsMADS51 transcript level was reduced
- OsGI, OsMADS51~OsMADS65, OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a, These results demonstrate that OsMADS51 functions downstream of OsGI
- OsGI, OsMADS51~OsMADS65, OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a, In summary, OsMADS51 is a novel flowering promoter that transmits a SD promotion signal from OsGI to Ehd1
- Ehd3, OsGI, Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date3, Transcript levels of OsGI, phytochrome genes, and Early heading date3 (Ehd3), which function upstream of Ghd7, were unchanged in the mutant
- Ghd7, OsGI, Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date3, Transcript levels of OsGI, phytochrome genes, and Early heading date3 (Ehd3), which function upstream of Ghd7, were unchanged in the mutant
- FKF1~OsFKF1, OsGI, Rice FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 OsFKF1 promotes flowering independent of photoperiod., Like Arabidopsis FKF1, which interacts with GI and CDF1, OsFKF1 also interacts with OsGI and OsCDF1 (also termed OsDOF12)
- OsGI, PHYA~OsPhyA, OsPhyA modulates rice flowering time mainly through OsGI under short days and Ghd7 under long days in the absence of phytochrome B., OsPhyA modulates rice flowering time mainly through OsGI under short days and Ghd7 under long days in the absence of phytochrome B.
- OsGI, PHYA~OsPhyA, OsPhyA modulates rice flowering time mainly through OsGI under short days and Ghd7 under long days in the absence of phytochrome B., These results indicated that OsPhyA influences flowering time mainly by affecting the expression of OsGI under SD and Ghd7 under LD when phytochrome B is absent
- OsAP37, OsGI, Mutation of OsGIGANTEA Leads to Enhanced Tolerance to Polyethylene Glycol-Generated Osmotic Stress in Rice., In addition, qRT-PCR and microarray analysis revealed that the transcript abundance of some osmotic stress response genes, such as OsDREB1E, OsAP37, OsAP59, OsLIP9, OsLEA3, OsRAB16A, and OsSalT, was significantly higher in osgi than in WT plants, suggesting that OsGI might be a negative regulator in the osmotic stress response in rice
- OsGI, OsLEA3~OsLEA3-1, Mutation of OsGIGANTEA Leads to Enhanced Tolerance to Polyethylene Glycol-Generated Osmotic Stress in Rice., In addition, qRT-PCR and microarray analysis revealed that the transcript abundance of some osmotic stress response genes, such as OsDREB1E, OsAP37, OsAP59, OsLIP9, OsLEA3, OsRAB16A, and OsSalT, was significantly higher in osgi than in WT plants, suggesting that OsGI might be a negative regulator in the osmotic stress response in rice
- OsGI, OsSalT, Mutation of OsGIGANTEA Leads to Enhanced Tolerance to Polyethylene Glycol-Generated Osmotic Stress in Rice., In addition, qRT-PCR and microarray analysis revealed that the transcript abundance of some osmotic stress response genes, such as OsDREB1E, OsAP37, OsAP59, OsLIP9, OsLEA3, OsRAB16A, and OsSalT, was significantly higher in osgi than in WT plants, suggesting that OsGI might be a negative regulator in the osmotic stress response in rice
- AP59~OsBIERF3~OsAP59, OsGI, Mutation of OsGIGANTEA Leads to Enhanced Tolerance to Polyethylene Glycol-Generated Osmotic Stress in Rice., In addition, qRT-PCR and microarray analysis revealed that the transcript abundance of some osmotic stress response genes, such as OsDREB1E, OsAP37, OsAP59, OsLIP9, OsLEA3, OsRAB16A, and OsSalT, was significantly higher in osgi than in WT plants, suggesting that OsGI might be a negative regulator in the osmotic stress response in rice
- OsDhn1~OsLip9, OsGI, Mutation of OsGIGANTEA Leads to Enhanced Tolerance to Polyethylene Glycol-Generated Osmotic Stress in Rice., In addition, qRT-PCR and microarray analysis revealed that the transcript abundance of some osmotic stress response genes, such as OsDREB1E, OsAP37, OsAP59, OsLIP9, OsLEA3, OsRAB16A, and OsSalT, was significantly higher in osgi than in WT plants, suggesting that OsGI might be a negative regulator in the osmotic stress response in rice
- OsDREB1E~OsDREB1-1~CR350, OsGI, Mutation of OsGIGANTEA Leads to Enhanced Tolerance to Polyethylene Glycol-Generated Osmotic Stress in Rice., In addition, qRT-PCR and microarray analysis revealed that the transcript abundance of some osmotic stress response genes, such as OsDREB1E, OsAP37, OsAP59, OsLIP9, OsLEA3, OsRAB16A, and OsSalT, was significantly higher in osgi than in WT plants, suggesting that OsGI might be a negative regulator in the osmotic stress response in rice
- OsGI, ROC4, GL2-type homeobox gene Roc4 in rice promotes flowering time preferentially under long days by repressing Ghd7, sativa GIGANTEA (OsGI) function upstream of Roc4
- OsGI, ROC4, GL2-type homeobox gene Roc4 in rice promotes flowering time preferentially under long days by repressing Ghd7, All of these findings are evidence that Roc4 is an LD-preferential flowering enhancer that functions downstream of phytochromes and OsGI, but upstream of Ghd7
- LTN1~OsPHO2, OsGI, Molecular interaction between PHO2 and GI in rice., Characterization of rice Osgi and Ospho2 mutants revealed that they displayed several similar phenotypic features supporting a physiological role for this interaction
- LTN1~OsPHO2, OsGI, Molecular interaction between PHO2 and GI in rice., Reduced growth, leaf tip necrosis, delayed flowering and over-accumulation of Pi in leaves compared to wild-type were shared features of Osgi and Ospho2 plants
- LTN1~OsPHO2, OsGI, Molecular interaction between PHO2 and GI in rice., Pi analysis of individual leaves demonstrated that Osgi, similar to Ospho2 mutants, were impaired in Pi remobilization from old to young leaves, albeit to a lesser extent
- LTN1~OsPHO2, OsGI, Molecular interaction between PHO2 and GI in rice., Transcriptome analyses revealed more than 55% of the genes differentially expressed in Osgi plants overlapped with the set of differentially expressed genes in Ospho2 plants
- LTN1~OsPHO2, OsGI, Molecular interaction between PHO2 and GI in rice., The interaction between OsPHO2 and OsGI links high-level regulators of Pi homeostasis and development in rice
- OsGI, OsMADS7~OsMADS45, Ectopic expression of OsMADS45 activates the upstream genes Hd3a and RFT1 at an early development stage causing early flowering in rice., OsMADS45 overexpression did not influence other floral regulator genes upstream of Hd1 and Ehd1, such as OsGI (OsGIGANTEA), Ehd2/Osld1/RID1 and OsMADS50