- Information
- Symbol: OsHKT1;5,SKC1,OsHKT8
- MSU: LOC_Os01g20160
- RAPdb: Os01g0307500
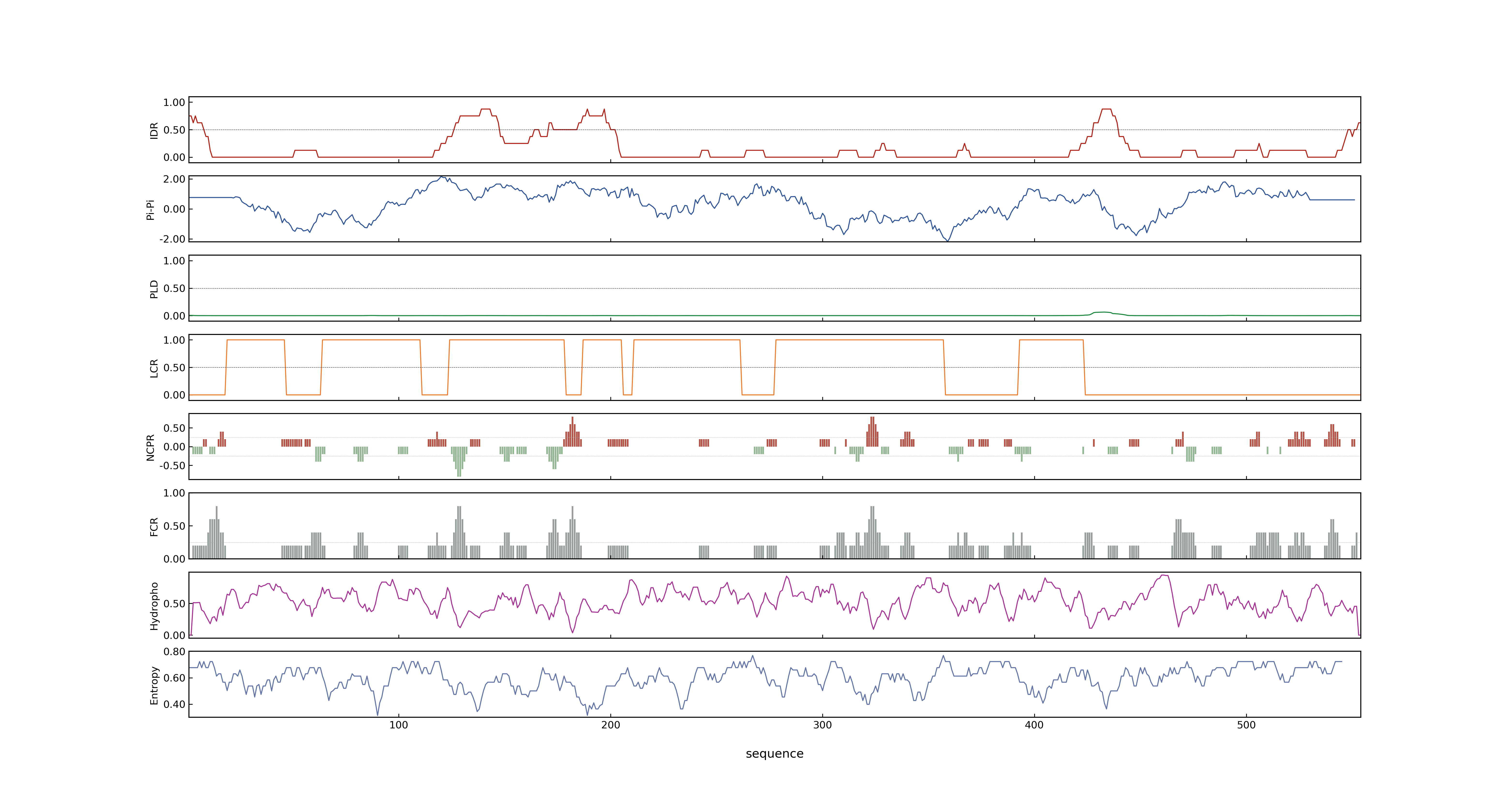
- PSP score
- LOC_Os01g20160.1: 0.5582
- PLAAC score
- LOC_Os01g20160.1: 0
- pLDDT score
- NA
- MolPhase score
- LOC_Os01g20160.1: 0.91880291
- MolPhase Result
- Publication
- QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance, 2004, Theor Appl Genet.
- Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice Oryza sativa L., 2012, BMC Plant Biol.
- Salinity tolerance, Na+ exclusion and allele mining of HKT1;5 in Oryza sativa and O. glaberrima: many sources, many genes, one mechanism?, 2013, BMC Plant Biol.
- A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress, 2010, Plant Cell Environ.
- A rice quantitative trait locus for salt tolerance encodes a sodium transporter, 2005, Nat Genet.
- A two-staged model of Na+ exclusion in rice explained by 3D modeling of HKT transporters and alternative splicing, 2012, PLoS One.
- OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice., 2017, Plant J.
- Genbank accession number
- Key message
- Here we present a review on vital physiological functions of HKT transporters including AtHKT1;1 and OsHKT1;5 in preventing shoot Na(+) over-accumulation by mediating Na(+) exclusion from xylem vessels in the presence of a large amount of Na(+) thereby protecting leaves from salinity stress
- We previously mapped a rice QTL, SKC1, that maintained K(+) homeostasis in the salt-tolerant variety under salt stress, consistent with the earlier finding that K(+) homeostasis is important in salt tolerance
- Physiological analysis suggested that SKC1 is involved in regulating K(+)/Na(+) homeostasis under salt stress, providing a potential tool for improving salt tolerance in crops
- SKC1 is preferentially expressed in the parenchyma cells surrounding the xylem vessels
- The association of leaf Na+ concentrations with cultivar-groups was very weak, but association with the OsHKT1;5 allele was generally strong
- Seven major and three minor alleles of OsHKT1;5 were identified, and their comparisons with the leaf Na+ concentration showed that the Aromatic allele conferred the highest exclusion and the Japonica allele the least
- In addition, lower expression of OsHKT1;5 and OsSOS1 in old leaves may decrease frequency of retrieving Na+ from old leaf cells
- To understand the molecular basis of this QTL, we isolated the SKC1 gene by map-based cloning and found that it encoded a member of HKT-type transporters
- Voltage-clamp analysis showed that SKC1 protein functions as a Na(+)-selective transporter
- For OsHKT1;5, both transcript abundance and protein structural features within the selectivity filter could control shoot Na(+) accumulation in a range of rice varieties
- OsHKT1;5 mediates Na(+) exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice.
- Additionally, direct introduction of (22) Na(+) tracer to leaf sheaths also demonstrated the involvement of OsHKT1;5 in xylem Na(+) unloading in leaf sheaths
- Together with the characteristic (22) Na(+) allocation in the blade of the developing immature leaf in the mutants, these results suggest a novel function of OsHKT1;5 in mediating Na(+) exclusion in the phloem to prevent Na(+) transfer to young leaf blades
- Our findings further demonstrate that the function of OsHKT1;5 is crucial over growth stages of rice, including the protection of the next generation seeds as well as of vital leaf blades under salt stress
- Immuno-staining using an anti-OsHKT1;5 peptide antibody indicated that OsHKT1;5 is localized in cells adjacent to the xylem in roots
- Furthermore, OsHKT1;5 was indicated to present in the plasma membrane and found to localize also in the phloem of diffuse vascular bundles in basal nodes
- Salt tolerance QTL analysis of rice has revealed that the SKC1 locus, which is involved in a higher K(+) /Na(+) ratio in shoots, corresponds to the OsHKT1;5 gene encoding a Na(+) -selective transporter
- However, physiological roles of OsHKT1;5 in rice exposed to salt stress remain elusive and no OsHKT1;5 gene disruption mutants have been characterized to date
- Salt stress-induced increases in the OsHKT1;5 transcript was observed in roots and basal stems including basal nodes
- Connection
- OsHKT1;5~SKC1~OsHKT8, OsNHA1~OsSOS1, Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice Oryza sativa L., In addition, lower expression of OsHKT1;5 and OsSOS1 in old leaves may decrease frequency of retrieving Na+ from old leaf cells
- OsHKT1;1~OsHKT4, OsHKT1;5~SKC1~OsHKT8, A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress, Here we present a review on vital physiological functions of HKT transporters including AtHKT1;1 and OsHKT1;5 in preventing shoot Na(+) over-accumulation by mediating Na(+) exclusion from xylem vessels in the presence of a large amount of Na(+) thereby protecting leaves from salinity stress
- OsHKT1;4, OsHKT1;5~SKC1~OsHKT8, A two-staged model of Na+ exclusion in rice explained by 3D modeling of HKT transporters and alternative splicing, Amongst these transporters, the cereal HKT1;4 and HKT1;5 are responsible for Na(+) exclusion from photosynthetic tissues, a key mechanism for plant salinity tolerance
- OsHKT1;5~SKC1~OsHKT8, OsPEX11, OsPEX11, a Peroxisomal Biogenesis Factor 11, Contributes to Salt Stress Tolerance in Oryza sativa., Furthermore, qPCR data suggested that OsPEX11 acted as a positive regulator of salt tolerance by reinforcing the expression of several well-known rice transporters (OsHKT2;1, OsHKT1;5, OsLti6a, OsLti6b, OsSOS1, OsNHX1, and OsAKT1) involved in Na(+)/K(+) homeostasis in transgenic plants under salinity
- OsHKT1;5~SKC1~OsHKT8, OsJRL, OsJRL, a rice jacalin-related mannose-binding lectin gene, enhances Escherichia coli viability under high-salinity stress and improves salinity tolerance of rice., Overexpression of OsJRL in rice also enhanced salinity tolerance and increased the expression levels of a number of stress-related genes, including three LEA (late embryogenesis abundant proteins) genes (OsLEA19a, OsLEA23 and OsLEA24), three Na(+) transporter genes (OsHKT1;3, OsHKT1;4 and OsHKT1;5) and two DREB genes (OsDREB1A and OsDREB2B)
- OsHKT1;5~SKC1~OsHKT8, OsMGT1, A magnesium transporter OsMGT1 plays a critical role in salt tolerance in rice., Here, we show evidence that a rice Mg transporter OsMGT1 is required for the salt-tolerance probably by regulating transport activity of OsHKT1;5, a key transporter for the removal of Na+ from the xylem sap at the root mature zone
- OsHKT1;5~SKC1~OsHKT8, OsMGT1, A magnesium transporter OsMGT1 plays a critical role in salt tolerance in rice., Furthermore, knockout of OsHKT1;5 in osmgt1 mutant background did not further increase its salt sensitivity
- OsHKT1;5~SKC1~OsHKT8, OsMGT1, A magnesium transporter OsMGT1 plays a critical role in salt tolerance in rice., Taken together, our results suggest that Mg2+ transported by OsMGT1 in the root mature zone is required for enhancing OsHKT1;5 activity, thereby restricting Na accumulation to the shoots
Prev Next