- Information
- Symbol: OsHOL2
- MSU: LOC_Os06g06040
- RAPdb: Os06g0153900
- PSP score
- LOC_Os06g06040.3: 0.0086
- LOC_Os06g06040.5: 0.028
- LOC_Os06g06040.1: 0.0649
- LOC_Os06g06040.2: 0.0246
- LOC_Os06g06040.4: 0.028
- LOC_Os06g06040.6: 0.028
- LOC_Os06g06040.7: 0.078
- PLAAC score
- LOC_Os06g06040.3: 0
- LOC_Os06g06040.5: 0
- LOC_Os06g06040.1: 0
- LOC_Os06g06040.2: 0
- LOC_Os06g06040.4: 0
- LOC_Os06g06040.6: 0
- LOC_Os06g06040.7: 0
- pLDDT score
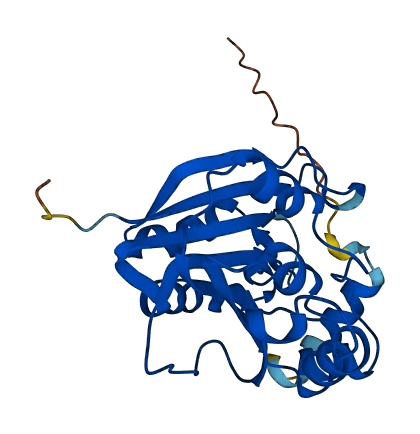
- 90.32
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os06g06040.1: 0.29248616
- LOC_Os06g06040.2: 0.34079569
- LOC_Os06g06040.3: 0.10356047
- LOC_Os06g06040.4: 0.38647326
- LOC_Os06g06040.5: 0.38647326
- LOC_Os06g06040.6: 0.38647326
- LOC_Os06g06040.7: 0.47192957
- MolPhase Result
- Publication
- Rice OsHOL1 and OsHOL2 proteins have S-adenosyl-L-methionine-dependent methyltransferase activities toward iodide ions, 2012, Plant Biotechnology.
- Arabidopsis HARMLESS TO OZONE LAYER Protein Methylates a Glucosinolate Breakdown Product and Functions in Resistance to Pseudomonas syringae pv. maculicola, 2009, Journal of Biological Chemistry.
- Genbank accession number
-
Key message
- Connection
- OsHOL1, OsHOL2, Arabidopsis HARMLESS TO OZONE LAYER Protein Methylates a Glucosinolate Breakdown Product and Functions in Resistance to Pseudomonas syringae pv. maculicola, Our previous studies demonstrated that AtHOL1 and its homologs, AtHOL2 and AtHOL3, have S-adenosyl-l-methionine-dependent methyltransferase activities
- OsHOL1, OsHOL2, Arabidopsis HARMLESS TO OZONE LAYER Protein Methylates a Glucosinolate Breakdown Product and Functions in Resistance to Pseudomonas syringae pv. maculicola, Analyses with the T-DNA insertion Arabidopsis mutants (hol1, hol2, and hol3) revealed that only hol1 showed increased sensitivity to NCS(-) in medium and a concomitant lack of CH(3)SCN synthesis upon tissue damage
Prev Next