- Information
- Symbol: OsLFL1
- MSU: LOC_Os01g51610
- RAPdb: Os01g0713600
- PSP score
- LOC_Os01g51610.1: 0.9703
- PLAAC score
- LOC_Os01g51610.1: 0
- pLDDT score
- 61.96
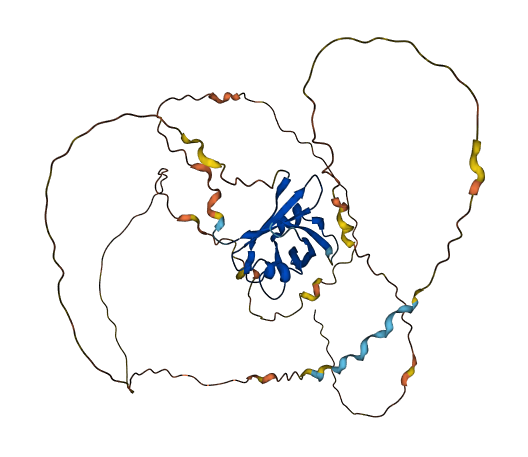
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os01g51610.1: 0.99994397
- MolPhase Result
- Publication
- Overexpression of transcription factor OsLFL1 delays flowering time in Oryza sativa, 2008, J Plant Physiol.
- Ectopic expression of OsLFL1 in rice represses Ehd1 by binding on its promoter, 2007, Biochem Biophys Res Commun.
- OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, 2009, Plant Cell Environ.
- OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice, 2013, Plant J.
- The rice GERMINATION DEFECTIVE 1, encoding a B3 domain transcriptional repressor, regulates seed germination and seedling development by integrating GA and carbohydrate metabolism, 2013, Plant J.
- Genbank accession number
- Key message
- Overexpression of transcription factor OsLFL1 delays flowering time in Oryza sativa
- In this study, it was found that the flowering promoting gene Ehd1 and its putative downstream genes were all repressed by OsLFL1
- In the mutant W378, the mutant gene coding OsLFL1, a putative B3 transcription factor gene, was ectopically expressed
- Furthermore, GD1 binds to the promoter of OsLFL1, a LEC2/FUS3-like gene of rice, via an RY element, leading to significant up-regulation of OsLFL1 and a large subset of seed maturation genes in the gd1 mutant
- As observed from osvil2, a null mutation of OsEMF2b caused late flowering by increasing OsLFL1 expression and decreasing Ehd1 expression
- Thus, we conclude that OsVIL2 functions together with PRC2 to induce flowering by repressing OsLFL1
- OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice
- These observations imply that OsMADS50 and OsMADS56 function antagonistically through OsLFL1-Ehd1 in regulating LD-dependent flowering
- Introduction of OsLFL1-RNAi into mutant W378 successfully down-regulated OsLFL1 expression and restored flowering to almost normal time, indicating that overexpression of OsLFL1 confers late flowering for mutant W378
- Thus, overexpression of OsLFL1 might delay the flowering of W378 by repressing the expression of Ehd1
- Connection
- Ehd1, OsLFL1, Ectopic expression of OsLFL1 in rice represses Ehd1 by binding on its promoter, In this study, it was found that the flowering promoting gene Ehd1 and its putative downstream genes were all repressed by OsLFL1
- Ehd1, OsLFL1, Ectopic expression of OsLFL1 in rice represses Ehd1 by binding on its promoter, Electrophoretic mobility shift assays (EMSA) and chromatin immunoprecipitation (ChIP) analyses suggest that OsLFL1 binds to the RY cis-elements (CATGCATG) in the promoter of the Ehd1 gene
- Ehd1, OsLFL1, Ectopic expression of OsLFL1 in rice represses Ehd1 by binding on its promoter, Thus, ectopically expressed OsLFL1 might repress Ehd1 via binding directly to the RY cis-elements in its promoter
- Ehd1, OsLFL1, Ectopic expression of OsLFL1 in rice represses Ehd1 by binding on its promoter, Ectopic expression of OsLFL1 in rice represses Ehd1 by binding on its promoter
- Ehd1, OsLFL1, OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, In the osmads50 mutants and 56OX transgenic plants, transcripts of Ehd1, Hd3a and RFT1 were reduced, although that of OsLFL1 increased
- Ehd1, OsLFL1, OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, These observations imply that OsMADS50 and OsMADS56 function antagonistically through OsLFL1-Ehd1 in regulating LD-dependent flowering
- Hd3a, OsLFL1, OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, In the osmads50 mutants and 56OX transgenic plants, transcripts of Ehd1, Hd3a and RFT1 were reduced, although that of OsLFL1 increased
- OsLFL1, OsMADS50~OsSOC1~DTH3, OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, In the osmads50 mutants and 56OX transgenic plants, transcripts of Ehd1, Hd3a and RFT1 were reduced, although that of OsLFL1 increased
- OsLFL1, OsMADS50~OsSOC1~DTH3, OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, These observations imply that OsMADS50 and OsMADS56 function antagonistically through OsLFL1-Ehd1 in regulating LD-dependent flowering
- OsLFL1, OsMADS56, OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, These observations imply that OsMADS50 and OsMADS56 function antagonistically through OsLFL1-Ehd1 in regulating LD-dependent flowering
- OsLFL1, RFT1, OsMADS50 and OsMADS56 function antagonistically in regulating long day LD-dependent flowering in rice, In the osmads50 mutants and 56OX transgenic plants, transcripts of Ehd1, Hd3a and RFT1 were reduced, although that of OsLFL1 increased
- Ehd1, OsLFL1, OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice, In osvil2 mutants OsLFL1 expression was increased, but that of Ehd1, Hd3a and RFT1 was reduced
- Ehd1, OsLFL1, OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice, As observed from osvil2, a null mutation of OsEMF2b caused late flowering by increasing OsLFL1 expression and decreasing Ehd1 expression
- Hd3a, OsLFL1, OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice, In osvil2 mutants OsLFL1 expression was increased, but that of Ehd1, Hd3a and RFT1 was reduced
- OsEMF2b, OsLFL1, OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice, As observed from osvil2, a null mutation of OsEMF2b caused late flowering by increasing OsLFL1 expression and decreasing Ehd1 expression
- OsLFL1, OsVIL2, OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice, In osvil2 mutants OsLFL1 expression was increased, but that of Ehd1, Hd3a and RFT1 was reduced
- OsLFL1, OsVIL2, OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice, Chromatin immunoprecipitation analyses showed that OsVIL2 was directly associated with OsLFL1 chromatin
- OsLFL1, OsVIL2, OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice, We also observed that H3K27me3 was significantly enriched by OsLFL1 chromatin in the wild type, but that this enrichment was diminished in the osvil2 mutants
- OsLFL1, OsVIL2, OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice, These results indicated that OsVIL2 epigenetically represses OsLFL1 expression
- OsLFL1, OsVIL2, OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice, As observed from osvil2, a null mutation of OsEMF2b caused late flowering by increasing OsLFL1 expression and decreasing Ehd1 expression
- OsLFL1, OsVIL2, OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice, Thus, we conclude that OsVIL2 functions together with PRC2 to induce flowering by repressing OsLFL1
- OsLFL1, OsVIL2, OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice, OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice
- OsLFL1, RFT1, OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice, In osvil2 mutants OsLFL1 expression was increased, but that of Ehd1, Hd3a and RFT1 was reduced
- GD1, OsLFL1, The rice GERMINATION DEFECTIVE 1, encoding a B3 domain transcriptional repressor, regulates seed germination and seedling development by integrating GA and carbohydrate metabolism, Furthermore, GD1 binds to the promoter of OsLFL1, a LEC2/FUS3-like gene of rice, via an RY element, leading to significant up-regulation of OsLFL1 and a large subset of seed maturation genes in the gd1 mutant
- GD1, OsLFL1, The rice GERMINATION DEFECTIVE 1, encoding a B3 domain transcriptional repressor, regulates seed germination and seedling development by integrating GA and carbohydrate metabolism, Plants over-expressing OsLFL1 partly mimic the gd1 mutant
Prev Next