- Information
- Symbol: OsMADS18
- MSU: LOC_Os07g41370
- RAPdb: Os07g0605200
- PSP score
- LOC_Os07g41370.1: 0.9143
- LOC_Os07g41370.2: 0.2647
- PLAAC score
- LOC_Os07g41370.1: 0
- LOC_Os07g41370.2: 0
- pLDDT score
- 78.99
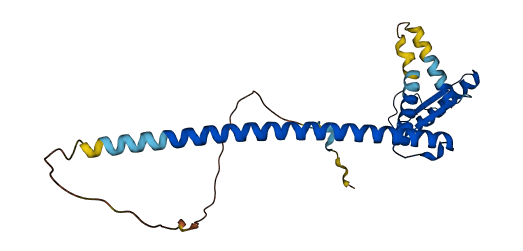
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os07g41370.1: 0.99992978
- LOC_Os07g41370.2: 0.99985516
- MolPhase Result
- Publication
- Ternary complex formation between MADS-box transcription factors and the histone fold protein NF-YB, 2002, J Biol Chem.
- Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 SOC1/AGL20 ortholog in rice, 2004, Plant J.
- Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene, 2012, Plant Cell.
- Functional characterization of OsMADS18, a member of the AP1/SQUA subfamily of MADS box genes, 2004, Plant Physiol.
- Genbank accession number
- Key message
- Overexpression of OsMADS18 in rice induced early flowering, and detailed histological analysis revealed that the formation of axillary shoot meristems was accelerated
- Silencing of OsMADS18 using an RNA interference approach did not result in any visible phenotypic alteration, indicating that OsMADS18 is probably redundant with other MADS box transcription factors
- A rice seed-specific NF-YB was identified as partner of OsMADS18 by two-hybrid screening
- RT-PCR analyses of the OsMADS50 KO and ubiquitin (ubi):OsMADS50 plants showed that OsMADS50 is an upstream regulator of OsMADS1, OsMADS14, OsMADS15, OsMADS18, and Hd (Heading date)3a, but works either parallel with or downstream of Hd1 and O
- Connection
- OsMADS18, OsNF-YB1, Ternary complex formation between MADS-box transcription factors and the histone fold protein NF-YB, OsMADS18, alone or in combination with a natural partner, interacts with OsNF-YB1 through the MADS and I regions
- OsMADS1~LHS1~AFO, OsMADS18, Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 SOC1/AGL20 ortholog in rice, RT-PCR analyses of the OsMADS50 KO and ubiquitin (ubi):OsMADS50 plants showed that OsMADS50 is an upstream regulator of OsMADS1, OsMADS14, OsMADS15, OsMADS18, and Hd (Heading date)3a, but works either parallel with or downstream of Hd1 and O
- OsMADS14, OsMADS18, Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 SOC1/AGL20 ortholog in rice, RT-PCR analyses of the OsMADS50 KO and ubiquitin (ubi):OsMADS50 plants showed that OsMADS50 is an upstream regulator of OsMADS1, OsMADS14, OsMADS15, OsMADS18, and Hd (Heading date)3a, but works either parallel with or downstream of Hd1 and O
- OsMADS15~DEP, OsMADS18, Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 SOC1/AGL20 ortholog in rice, RT-PCR analyses of the OsMADS50 KO and ubiquitin (ubi):OsMADS50 plants showed that OsMADS50 is an upstream regulator of OsMADS1, OsMADS14, OsMADS15, OsMADS18, and Hd (Heading date)3a, but works either parallel with or downstream of Hd1 and O
- OsMADS18, OsMADS50~OsSOC1~DTH3, Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 SOC1/AGL20 ortholog in rice, RT-PCR analyses of the OsMADS50 KO and ubiquitin (ubi):OsMADS50 plants showed that OsMADS50 is an upstream regulator of OsMADS1, OsMADS14, OsMADS15, OsMADS18, and Hd (Heading date)3a, but works either parallel with or downstream of Hd1 and O
- Hd1, OsMADS18, Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 SOC1/AGL20 ortholog in rice, RT-PCR analyses of the OsMADS50 KO and ubiquitin (ubi):OsMADS50 plants showed that OsMADS50 is an upstream regulator of OsMADS1, OsMADS14, OsMADS15, OsMADS18, and Hd (Heading date)3a, but works either parallel with or downstream of Hd1 and O
- OsMADS14, OsMADS18, Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene, Using laser microdissection and microarrays, we found that expression of PANICLE PHYTOMER2 (PAP2) and three APETALA1 (AP1)/FRUITFULL (FUL)-like genes (MADS14, MADS15, and MADS18) is induced in the SAM during meristem phase transition in rice (Oryza sativa)
- OsMADS15~DEP, OsMADS18, Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene, Using laser microdissection and microarrays, we found that expression of PANICLE PHYTOMER2 (PAP2) and three APETALA1 (AP1)/FRUITFULL (FUL)-like genes (MADS14, MADS15, and MADS18) is induced in the SAM during meristem phase transition in rice (Oryza sativa)
- OsMADS18, OsMADS34~PAP2, Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene, Using laser microdissection and microarrays, we found that expression of PANICLE PHYTOMER2 (PAP2) and three APETALA1 (AP1)/FRUITFULL (FUL)-like genes (MADS14, MADS15, and MADS18) is induced in the SAM during meristem phase transition in rice (Oryza sativa)
- OsMADS18, OsMADS7~OsMADS45, Ectopic expression of OsMADS45 activates the upstream genes Hd3a and RFT1 at an early development stage causing early flowering in rice., These results indicate that in transgenic rice, OsMADS45 overexpressing ectopically activates the upstream genes Hd3a and RFT1 at early development stage and up-regulates the expression of OsMADS14 and OsMADS18, which induces early flowering
Prev Next