- Information
- Symbol: OsMADS2
- MSU: LOC_Os01g66030
- RAPdb: Os01g0883100
- PSP score
- LOC_Os01g66030.1: 0.0057
- LOC_Os01g66030.2: 0.0124
- PLAAC score
- LOC_Os01g66030.1: 0
- LOC_Os01g66030.2: 0
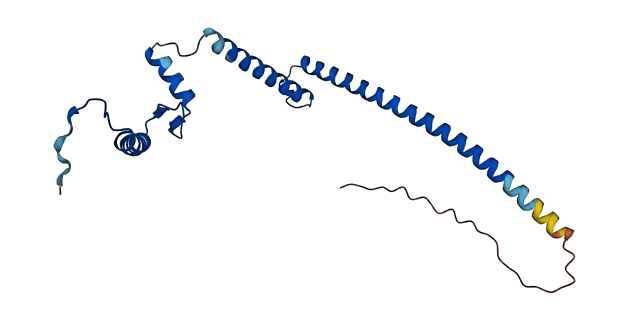
- pLDDT score
- 86.08
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os01g66030.1: 0.02268886
- LOC_Os01g66030.2: 0.00305004
- MolPhase Result
- Publication
- Double-stranded RNA interference of a rice PI/GLO paralog, OsMADS2, uncovers its second-whorl-specific function in floral organ patterning, 2003, Genetics.
- MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress, 2007, BMC Genomics.
- Divergent regulatory OsMADS2 functions control size, shape and differentiation of the highly derived rice floret second-whorl organ, 2007, Genetics.
- Unequal genetic redundancy of rice PISTILLATA orthologs, OsMADS2 and OsMADS4, in lodicule and stamen development, 2008, Plant Cell Physiol.
- The superwoman1-cleistogamy2 mutant is a novel resource for gene containment in rice., 2016, Plant Biotechnol J.
- Genbank accession number
- Key message
- We ascribe a function for OsMADS2 in controlling cell division and differentiation along the proximal-distal axis
- The global architecture of transcripts regulated by OsMADS2 gives insights into the regulation of cell division and vascular differentiation that together can form this highly modified grass organ with important functions in floret opening and stamen emergence independent of the paralogous gene OsMADS4
- OsMADS2 is required to trigger parenchymatous and lodicule-specific vascular development while maintaining a small organ size
- Our data implicate the developmentally late spatially restricted accumulation of OsMADS2 transcripts in the differentiating lodicule to control growth of these regions
- Grasses have duplicated PI/GLO-like genes and in rice (Oryza sativa) one these genes, OsMADS2, controls lodicule formation without affecting stamen development
- Double-stranded RNA interference of a rice PI/GLO paralog, OsMADS2, uncovers its second-whorl-specific function in floral organ patterning
- Unequal genetic redundancy of rice PISTILLATA orthologs, OsMADS2 and OsMADS4, in lodicule and stamen development
- Lodicule identity and development are specified by the action of protein complexes involving the SPW1 and OsMADS2 transcription factors
- Connection
- OsMADS2, OsMADS4, Divergent regulatory OsMADS2 functions control size, shape and differentiation of the highly derived rice floret second-whorl organ, The global architecture of transcripts regulated by OsMADS2 gives insights into the regulation of cell division and vascular differentiation that together can form this highly modified grass organ with important functions in floret opening and stamen emergence independent of the paralogous gene OsMADS4
- OsMADS2, OsMADS4, Unequal genetic redundancy of rice PISTILLATA orthologs, OsMADS2 and OsMADS4, in lodicule and stamen development, Two homologs of PISTILLATA have been identified in rice: OsMADS2 and OsMADS4
- OsMADS2, OsMADS4, Unequal genetic redundancy of rice PISTILLATA orthologs, OsMADS2 and OsMADS4, in lodicule and stamen development, Although OsMADS2 plays an important role in lodicule development, OsMADS4 also supports the specification of lodicule identity
- OsMADS2, OsMADS4, Unequal genetic redundancy of rice PISTILLATA orthologs, OsMADS2 and OsMADS4, in lodicule and stamen development, Consistent with their redundant functions, both OsMADS2 and OsMADS4 interact with the unique rice AP3 ortholog SPW1
- OsMADS2, OsMADS4, Unequal genetic redundancy of rice PISTILLATA orthologs, OsMADS2 and OsMADS4, in lodicule and stamen development, Unequal genetic redundancy of rice PISTILLATA orthologs, OsMADS2 and OsMADS4, in lodicule and stamen development
- CFO1~OsMADS32, OsMADS2, OsMADS32 interacts with PI-like proteins and regulates rice flower development., By yeast two hybrid and bimolecular fluorescence complementation assays, we revealed that the insertion of eight amino acids or deletion of the internal region in the K1 subdomain of OsMADS32 affects the interaction between OsMADS32 with PISTILLATA (PI)-like proteins OsMADS2 and OsMADS4
- OsMADS16~SPW1, OsMADS2, The superwoman1-cleistogamy2 mutant is a novel resource for gene containment in rice., Lodicule identity and development are specified by the action of protein complexes involving the SPW1 and OsMADS2 transcription factors
- OsMADS16~SPW1, OsMADS2, The superwoman1-cleistogamy2 mutant is a novel resource for gene containment in rice., In the superwoman1-cleistogamy1 (spw1-cls1) mutant, SPW1 is impaired for heterodimerization with OsMADS2 and consequently spw1-cls1 shows thin, ineffective lodicules
Prev Next