- Information
- Symbol: OsMADS34,PAP2
- MSU: LOC_Os03g54170
- RAPdb: Os03g0753100
- PSP score
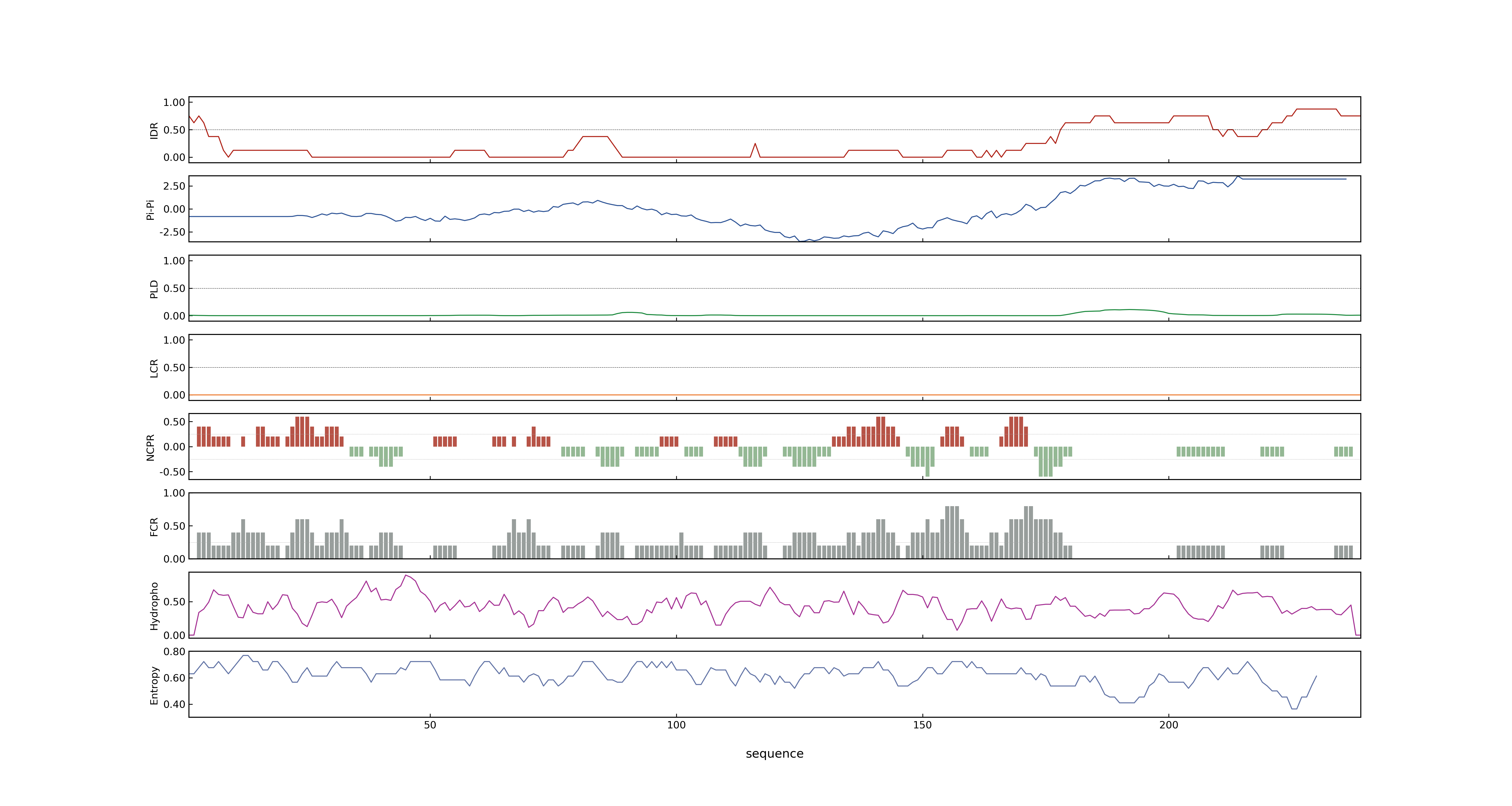
- LOC_Os03g54170.1: 0.1234
- PLAAC score
- LOC_Os03g54170.1: 0
- pLDDT score
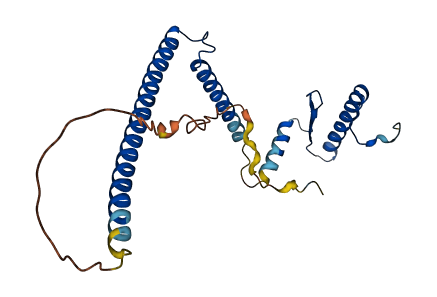
- 79.93
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os03g54170.1: 0.96967756
- MolPhase Result
- Publication
- [The pleiotropic SEPALLATA-like gene OsMADS34 reveals that the ‘empty glumes’ of rice Oryza sativa spikelets are in fact rudimentary lemmas](http://www.ncbi.nlm.nih.gov/pubmed?term=The pleiotropic SEPALLATA-like gene OsMADS34 reveals that the ‘empty glumes’ of rice Oryza sativa spikelets are in fact rudimentary lemmas%5BTitle%5D), 2014, New Phytol.
- PANICLE PHYTOMER2 PAP2, encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice, 2010, Plant Cell Physiol.
- Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene, 2012, Plant Cell.
- The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development, 2010, Plant Physiol.
- Regulatory Role of OsMADS34 in the Determination of Glumes Fate, Grain Yield, and Quality in Rice., 2016, Front Plant Sci.
- Regulatory network and genetic interactions established by OsMADS34 in rice inflorescence and spikelet morphogenesis., 2017, J Integr Plant Biol.
-
Genbank accession number
- Key message
- Using laser microdissection and microarrays, we found that expression of PANICLE PHYTOMER2 (PAP2) and three APETALA1 (AP1)/FRUITFULL (FUL)-like genes (MADS14, MADS15, and MADS18) is induced in the SAM during meristem phase transition in rice (Oryza sativa)
- In addition, osmads34 mutants displayed a decreased spikelet number and altered spikelet morphology, with lemma/leaf-like elongated sterile lemmas
- We demonstrate the ubiquitous expression of OsMADS34 in roots, leaves, and primordia of inflorescence and spikelet organs
- Moreover, analysis of the double mutant osmads34 osmads1 suggests that OsMADS34 specifies the identities of floral organs, including the lemma/palea, lodicules, stamens, and carpel, in combination with another rice SEP-like gene, OsMADS1
- Furthermore, the precocious flowering phenotype caused by the overexpression of Hd3a, a rice florigen gene, was weakened in pap2-1 mutants
- In the panicle of the panicle phytomer2-1 (pap2-1) mutant, the pattern of meristem initiation is disorganized and newly formed meristems show reduced competency to become spikelet meristems, resulting in the transformation of early arising spikelets into rachis branches
- PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice
- Here, we report the biological role of one SEPALLATA (SEP)-like gene, OsMADS34, in controlling the development of inflorescences and spikelets in rice (Oryza sativa)
- Compared with the wild type, osmads34 mutants developed altered inflorescence morphology, with an increased number of primary branches and a decreased number of secondary branches
- Collectively, our study suggests that the origin and diversification of OsMADS34 and OsMADS1 contribute to the origin of distinct grass inflorescences and spikelets
- The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development
- Our findings support the hypothesis that OUIs originated from the lemmas of degenerate florets under the negative control of OsMADS34
- The pleiotropic SEPALLATA-like gene OsMADS34 reveals that the ‘empty glumes’ of rice (Oryza sativa) spikelets are in fact rudimentary lemmas
- We propose that PAP2 is a positive regulator of spikelet meristem identity
- PAP2 expression starts the earliest among the five SEP genes, and a low but significant level of PAP2 mRNA was detected in the inflorescence meristem, in branch meristems immediately after the transition, and in glume primordia, consistent with its role in the early development of spikelet formation
- Here we present a novel mutant of the rice SEPALLATA-like gene OsMADS34 which develops, in addition to disorganized branches and sterile seeds, elongated OUIs
- Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene
- Further depletion of PAP2 function from these triple knockdown plants inhibited the transition of the meristem to the IM
- Based on these results, we propose that PAP2 and the three AP1/FUL-like genes coordinately act in the meristem to specify the identity of the IM downstream of the florigen signal
- Regulatory Role of OsMADS34 in the Determination of Glumes Fate, Grain Yield, and Quality in Rice.
- In this study, we used m34-z, a new mutant allele of the rice (Oryza sativa) E-class gene OsMADS34, to examine OsMADS34 function in determining the identities of glumes (rudimentary glume and sterile lemma) and grain size
- Interesting, transcriptional activity analysis revealed that OsMADS34 protein was a transcription repressor and it may influence grain yield by suppressing the expressions of BG1, GW8, GW2, and GL7 in the m34-z mutant
- These findings revealed that OsMADS34 largely affects grain yield by affecting the size of grains from the secondary branches
- In the m34-z mutant, both the rudimentary glume and sterile lemma were homeotically converted to the lemma-like organ and acquired the lemma identity, suggesting that OsMADS34 plays important roles in the development of glumes
- Regulatory network and genetic interactions established by OsMADS34 in rice inflorescence and spikelet morphogenesis.
- Collectively, these findings provide insights into the regulatory function of OsMADS34 in rice inflorescence and spikelet development
- longer and wider sterile lemmas that were converted into lemma/palea like organs, suggesting that ELE and OsMADS34 synergistically control the sterile lemma development
- OsMADS34 may act together OsMADS15 in controlling sterile lemma development
- Genetic analysis revealed that OsMADS34 controls different aspects of inflorescence structure, branching and meristem activity synergistically with LAX PANICLE1 (LAX1) and FLORAL ORGAN NUMBER4 (FON4), as evidenced by the enhanced phenotypes of osmads34 lax1 and osmads34 fon4 compared with the single mutants
- Microarray analysis showed that, at the very early stages of inflorescence formation, dysfunction of OsMADS34 caused altered expression of 379 genes that are associated with protein modification and degradation, transcriptional regulation, signaling and metabolism activity
- Additionally, double mutant between osmads34 and the sterile lemma defective mutant elongated empty glume (ele) displayed an enhanced phenotype, i
- Connection
- Hd3a, OsMADS34~PAP2, Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene, Furthermore, the precocious flowering phenotype caused by the overexpression of Hd3a, a rice florigen gene, was weakened in pap2-1 mutants
- OsMADS14, OsMADS34~PAP2, Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene, Using laser microdissection and microarrays, we found that expression of PANICLE PHYTOMER2 (PAP2) and three APETALA1 (AP1)/FRUITFULL (FUL)-like genes (MADS14, MADS15, and MADS18) is induced in the SAM during meristem phase transition in rice (Oryza sativa)
- OsMADS15~DEP, OsMADS34~PAP2, Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene, Further depletion of PAP2 function from these triple knockdown plants inhibited the transition of the meristem to the IM
- OsMADS15~DEP, OsMADS34~PAP2, Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene, Using laser microdissection and microarrays, we found that expression of PANICLE PHYTOMER2 (PAP2) and three APETALA1 (AP1)/FRUITFULL (FUL)-like genes (MADS14, MADS15, and MADS18) is induced in the SAM during meristem phase transition in rice (Oryza sativa)
- OsMADS15~DEP, OsMADS34~PAP2, Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene, PAP2 physically interacts with MAD14 and MADS15 in vivo
- OsMADS18, OsMADS34~PAP2, Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene, Using laser microdissection and microarrays, we found that expression of PANICLE PHYTOMER2 (PAP2) and three APETALA1 (AP1)/FRUITFULL (FUL)-like genes (MADS14, MADS15, and MADS18) is induced in the SAM during meristem phase transition in rice (Oryza sativa)
- OsMADS1~LHS1~AFO, OsMADS34~PAP2, The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development, Moreover, analysis of the double mutant osmads34 osmads1 suggests that OsMADS34 specifies the identities of floral organs, including the lemma/palea, lodicules, stamens, and carpel, in combination with another rice SEP-like gene, OsMADS1
- OsMADS1~LHS1~AFO, OsMADS34~PAP2, The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development, Collectively, our study suggests that the origin and diversification of OsMADS34 and OsMADS1 contribute to the origin of distinct grass inflorescences and spikelets
- OsEMF2b, OsMADS34~PAP2, The polycomb group gene EMF2B is essential for maintenance of floral meristem determinacy in rice, Transcriptome analysis identified the E-function genes OsMADS1, OsMADS6 and OsMADS34 as differentially expressed in the emf2b mutant compared with wild type.
- OsEMF2b, OsMADS34~PAP2, The polycomb group gene EMF2B is essential for maintenance of floral meristem determinacy in rice, OsMADS1 and OsMADS6, known to be required for meristem determinacy in rice, have reduced expression in the emf2b mutant, whereas OsMADS34 which interacts genetically with OsMADS1 was ectopically expressed.
- BG1, OsMADS34~PAP2, Regulatory Role of OsMADS34 in the Determination of Glumes Fate, Grain Yield, and Quality in Rice., Interesting, transcriptional activity analysis revealed that OsMADS34 protein was a transcription repressor and it may influence grain yield by suppressing the expressions of BG1, GW8, GW2, and GL7 in the m34-z mutant
- GW2, OsMADS34~PAP2, Regulatory Role of OsMADS34 in the Determination of Glumes Fate, Grain Yield, and Quality in Rice., Interesting, transcriptional activity analysis revealed that OsMADS34 protein was a transcription repressor and it may influence grain yield by suppressing the expressions of BG1, GW8, GW2, and GL7 in the m34-z mutant
- GW7~GL7~SLG7, OsMADS34~PAP2, Regulatory Role of OsMADS34 in the Determination of Glumes Fate, Grain Yield, and Quality in Rice., Interesting, transcriptional activity analysis revealed that OsMADS34 protein was a transcription repressor and it may influence grain yield by suppressing the expressions of BG1, GW8, GW2, and GL7 in the m34-z mutant
- OsMADS34~PAP2, qGW8~OsSPL16~GW8, Regulatory Role of OsMADS34 in the Determination of Glumes Fate, Grain Yield, and Quality in Rice., Interesting, transcriptional activity analysis revealed that OsMADS34 protein was a transcription repressor and it may influence grain yield by suppressing the expressions of BG1, GW8, GW2, and GL7 in the m34-z mutant
- FON2~FON4~TG1, OsMADS34~PAP2, Regulatory network and genetic interactions established by OsMADS34 in rice inflorescence and spikelet morphogenesis., Genetic analysis revealed that OsMADS34 controls different aspects of inflorescence structure, branching and meristem activity synergistically with LAX PANICLE1 (LAX1) and FLORAL ORGAN NUMBER4 (FON4), as evidenced by the enhanced phenotypes of osmads34 lax1 and osmads34 fon4 compared with the single mutants
- LAX1, OsMADS34~PAP2, Regulatory network and genetic interactions established by OsMADS34 in rice inflorescence and spikelet morphogenesis., Genetic analysis revealed that OsMADS34 controls different aspects of inflorescence structure, branching and meristem activity synergistically with LAX PANICLE1 (LAX1) and FLORAL ORGAN NUMBER4 (FON4), as evidenced by the enhanced phenotypes of osmads34 lax1 and osmads34 fon4 compared with the single mutants
- OsMADS15~DEP, OsMADS34~PAP2, Regulatory network and genetic interactions established by OsMADS34 in rice inflorescence and spikelet morphogenesis., OsMADS34 may act together OsMADS15 in controlling sterile lemma development
Prev Next