- Information
- Symbol: OsMYB103L,CEF1
- MSU: LOC_Os08g05520
- RAPdb: Os08g0151300
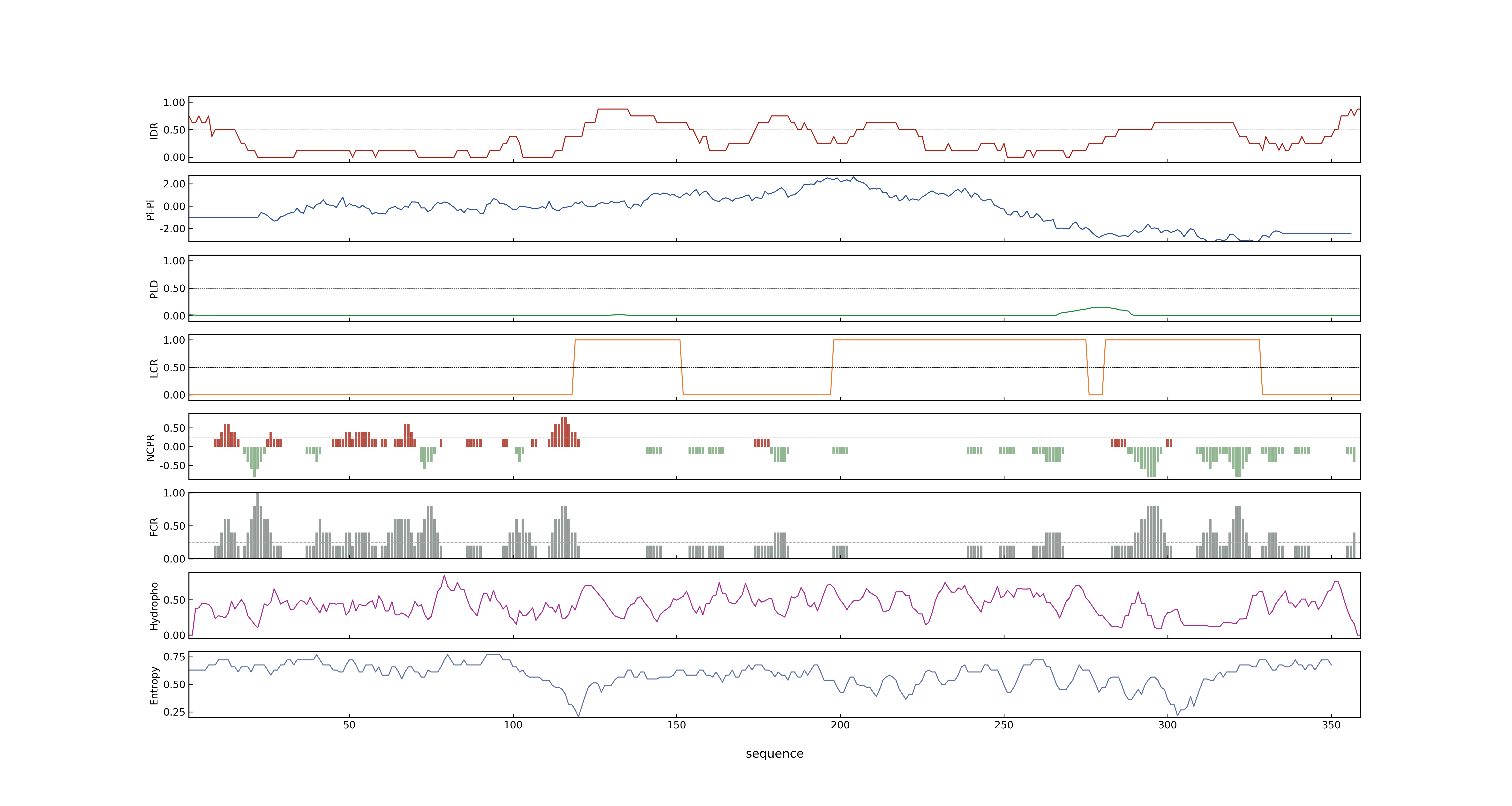
- PSP score
- LOC_Os08g05520.1: 0.8752
- PLAAC score
- LOC_Os08g05520.1: 0
- pLDDT score
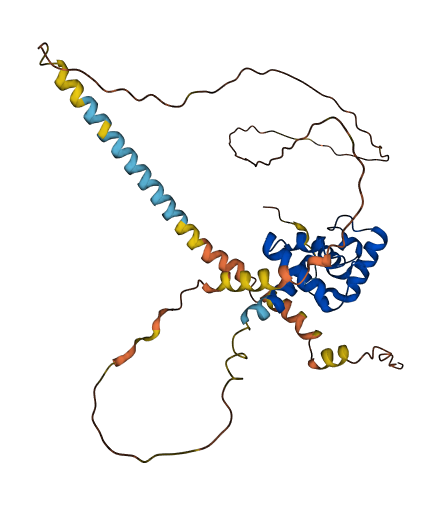
- 63.64
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os08g05520.1: 0.99612075
- MolPhase Result
- Publication
-
Genbank accession number
- Key message
- OsMYB103L, an R2R3-MYB transcription factor, influences leaf rolling and mechanical strength in rice (Oryza sativa L.).
- Leaf rolling is one of the important traits in rice (Oryza sativa L
- Overexpression of OsMYB103L in rice resulted in a rolled leaf phenotype
- These findings suggest that OsMYB103L may target CESA genes for regulation of cellulose synthesis and could potentially be engineered for desirable leaf shape and mechanical strength in rice
- MYB transcription factors are one of the largest gene families and have important roles in plant development, metabolism and stress responses
- In this study, we report the functional characterization of a rice gene, OsMYB103L, which encodes an R2R3-MYB transcription factor
- Further analyses showed that expression levels of several cellulose synthase genes (CESAs) were significantly increased, as was the cellulose content in OsMYB103L overexpressing lines
- Knockdown of OsMYB103L by RNA interference led to a decreased level of cellulose content and reduced mechanical strength in leaves
- ) breeding
- OsMYB103L may also function as a master switch to regulate the expression of several downstream TFs, which involved in secondary cell wall biosynthesis
- Our findings revealed that OsMYB103L plays an important role in GA-regulating secondary cell wall synthesis, and the manipulation of this gene provide a new strategy to help the straw decay in soil
- Furthermore, OsMYB103L physically interacts with SLENDER RICE1 (SLR1), a DELLA repressor of GA signaling, and involved in GA-mediated regulation of cellulose synthesis pathway
- OsMYB103L mediates cellulose biosynthesis and secondary walls formation mainly through directly binding the CESA4, CESA7, CESA9 and BC1 promoters and regulating their expression
- Biochemical assays demonstrated that OsMYB103L is a nuclear protein and shows high transcriptional activation activity at C-terminus
- Connection
- BC1, OsMYB103L~CEF1, CEF1/OsMYB103L is involved in GA-mediated regulation of secondary wall biosynthesis in rice., OsMYB103L mediates cellulose biosynthesis and secondary walls formation mainly through directly binding the CESA4, CESA7, CESA9 and BC1 promoters and regulating their expression
- OsMYB103L~CEF1, SLR1~OsGAI, CEF1/OsMYB103L is involved in GA-mediated regulation of secondary wall biosynthesis in rice., Furthermore, OsMYB103L physically interacts with SLENDER RICE1 (SLR1), a DELLA repressor of GA signaling, and involved in GA-mediated regulation of cellulose synthesis pathway
- Bc6~OsCesA9, OsMYB103L~CEF1, CEF1/OsMYB103L is involved in GA-mediated regulation of secondary wall biosynthesis in rice, OsMYB103L mediates cellulose biosynthesis and secondary walls formation mainly through directly binding the CESA4, CESA7, CESA9 and BC1 promoters and regulating their expression
- OsCesA4~Bc7~bc11, OsMYB103L~CEF1, CEF1/OsMYB103L is involved in GA-mediated regulation of secondary wall biosynthesis in rice, OsMYB103L mediates cellulose biosynthesis and secondary walls formation mainly through directly binding the CESA4, CESA7, CESA9 and BC1 promoters and regulating their expression
- OsCesA7, OsMYB103L~CEF1, CEF1/OsMYB103L is involved in GA-mediated regulation of secondary wall biosynthesis in rice, OsMYB103L mediates cellulose biosynthesis and secondary walls formation mainly through directly binding the CESA4, CESA7, CESA9 and BC1 promoters and regulating their expression
Prev Next