- Information
- Symbol: OsNAC5
- MSU: LOC_Os11g08210
- RAPdb: Os11g0184900
- PSP score
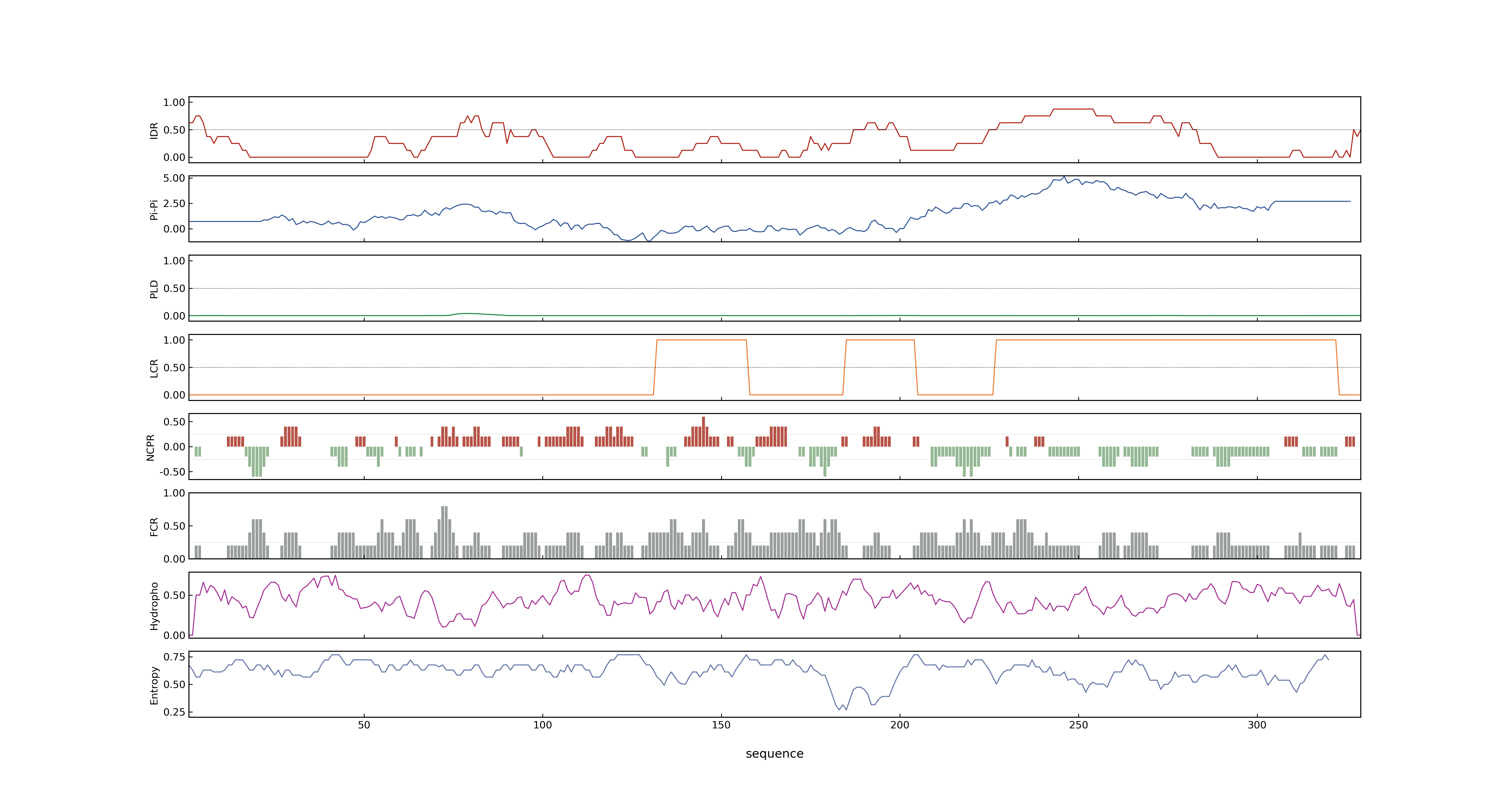
- LOC_Os11g08210.1: 0.1036
- PLAAC score
- LOC_Os11g08210.1: 0
- pLDDT score
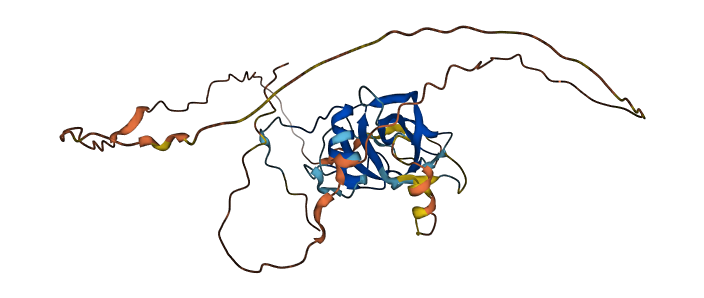
- 64.47
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os11g08210.1: 0.99693858
- MolPhase Result
- Publication
- Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress, 2011, Planta.
- The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice, 2010, Mol Genet Genomics.
- OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field, 2013, Plant Biotechnol J.
- Molecular analysis of the NAC gene family in rice, 2000, Mol Gen Genet.
- Identification of up-regulated genes in flag leaves during rice grain filling and characterization of OsNAC5, a new ABA-dependent transcription factor, 2009, Planta.
- Genbank accession number
- Key message
- To understand the functions of transcription factor OsNAC5 in response to abiotic stress, we generated transgenic rice plants with knockdown OsNAC5 by RNA-interfered (RNAi) and overexpressing OsNAC5, and investigated the effects of cold, drought and salt stress on wild-type (WT), RNAi and overexpression rice lines
- These findings highlight the important role of OsNAC5 played in the tolerance of rice plants to abiotic stress by regulating downstream targets associated with accumulation of compatible solutes, Na(+) ions, H(2)O(2) and malondialdehyde
- Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress
- Both the RCc3:OsNAC5 and GOS2:OsNAC5 plants were found to have larger roots due to an enlarged stele and aerenchyma at flowering stage
- Field evaluations over three growing seasons revealed that the grain yield of the RCc3:OsNAC5 and GOS2:OsNAC5 plants were increased by 9%-23% and 9%-26% under normal conditions, respectively
- Under drought conditions, however, RCc3:OsNAC5 plants showed a significantly higher grain yield of 22%-63%, whilst the GOS2:OsNAC5 plants showed a reduced or similar yield to the nontransgenic (NT) controls
- Our present findings demonstrate that the root-specific overexpression of OsNAC5 enlarges roots significantly and thereby enhances drought tolerance and grain yield under field conditions
- OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field
- The root diameter was enlarged to a greater extent in the RCc3:OsNAC5, suggesting the importance of this phenotype for enhanced drought tolerance
- Of the genes specifically up-regulated in the RCc3:OsNAC5 roots, GLP, PDX, MERI5 and O-methyltransferase were implicated in root growth and development
- Expression of OsNAC5 is induced by abiotic stresses such as drought, cold, high salinity, abscisic acid and methyl jasmonic acid
- The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice
- We show that OsNAC5 expression is up-regulated by natural (aging) and induced senescence processes (dark, ABA application, high salinity, cold and Fe-deficiency) and its expression is not affected in the presence of 6-benzylaminopurine (a senescence inhibitor) under dark-induced senescence
- Using four different rice cultivars, we show that OsNAC5 up-regulation is higher and earlier in flag leaves and panicles of IR75862 plants, which have higher seed concentrations of Fe, Zn and protein
- We suggest that OsNAC5 is a novel senescence-associated ABA-dependent NAC transcription factor and its function could be related to Fe, Zn and amino acids remobilization from green tissues to seeds
- The transcription factor OsNAC5 in rice is a member of the plant-specific NAC family that regulates stress responses
- OsNAC5-overexpressing transgenic plants also had improved tolerance to high salinity compared to control plants
- Salt induction of OsNAC5 expression is abolished by nicotinamide, an inhibitor of ABA effects
- Collectively, our results indicate that the stress-responsive proteins OsNAC5 and OsNAC6 are transcriptional activators that enhance stress tolerance by upregulating the expression of stress-inducible rice genes such as OsLEA3, although the effects of these proteins on growth are different
- Furthermore, because OsNAC5 overexpression did not retard growth, OsNAC5 may be a useful gene that can improve the stress tolerance of rice without affecting its growth
- Differential expression of selected genes (encoding 7 transport proteins, the OsNAS3 enzyme and the OsNAC5 transcription factor) was confirmed by quantitative RT-PCR
- Identification of up-regulated genes in flag leaves during rice grain filling and characterization of OsNAC5, a new ABA-dependent transcription factor
- In addition, knockdown and overexpression of OsNAC5 enhanced and reduced accumulation of malondialdehyde and H(2)O(2), suggesting that knockdown of OsNAC5 renders RNAi plants more susceptible to oxidative damage
- Here, we report the results of field evaluations of transgenic rice plants overexpressing OsNAC5, under the control of either the root-specific (RCc3) or constitutive (GOS2) promoters
- Also identified were 19 and 18 up-regulated genes that are specific to the RCc3:OsNAC5 and GOS2:OsNAC5 roots, respectively
- Connection
- OsLEA3~OsLEA3-1, OsNAC5, The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice, By microarray analysis, many stress-inducible genes, including the “late embryogenesis abundant” gene OsLEA3, were upregulated in rice plants that overexpressed OsNAC5
- OsLEA3~OsLEA3-1, OsNAC5, The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice, By gel mobility shift assay, OsNAC5 and OsNAC6 were shown to bind to the OsLEA3 promoter
- OsLEA3~OsLEA3-1, OsNAC5, The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice, Collectively, our results indicate that the stress-responsive proteins OsNAC5 and OsNAC6 are transcriptional activators that enhance stress tolerance by upregulating the expression of stress-inducible rice genes such as OsLEA3, although the effects of these proteins on growth are different
- OsNAC19~SNAC1~OsNAC9, OsNAC5, The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice, Pull-down assays revealed that OsNAC5 interacts with OsNAC5, OsNAC6 and SNAC1
- OsNAC5, OsNAC6~SNAC2, The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice, Pull-down assays revealed that OsNAC5 interacts with OsNAC5, OsNAC6 and SNAC1
- OsNAC5, OsNAC6~SNAC2, The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice, By gel mobility shift assay, OsNAC5 and OsNAC6 were shown to bind to the OsLEA3 promoter
- OsNAC5, OsNAC6~SNAC2, The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice, Collectively, our results indicate that the stress-responsive proteins OsNAC5 and OsNAC6 are transcriptional activators that enhance stress tolerance by upregulating the expression of stress-inducible rice genes such as OsLEA3, although the effects of these proteins on growth are different
- OsNAC5, OsNAS3, Identification of up-regulated genes in flag leaves during rice grain filling and characterization of OsNAC5, a new ABA-dependent transcription factor, Differential expression of selected genes (encoding 7 transport proteins, the OsNAS3 enzyme and the OsNAC5 transcription factor) was confirmed by quantitative RT-PCR
- ONAC106, OsNAC5, Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle., Using yeast one-hybrid assays, we found that ONAC106 binds to the promoter regions of SGR, NYC1, OsNAC5, and LPA1
Prev Next