- Information
- Symbol: OsNAP
- MSU: LOC_Os03g21060
- RAPdb: Os03g0327800
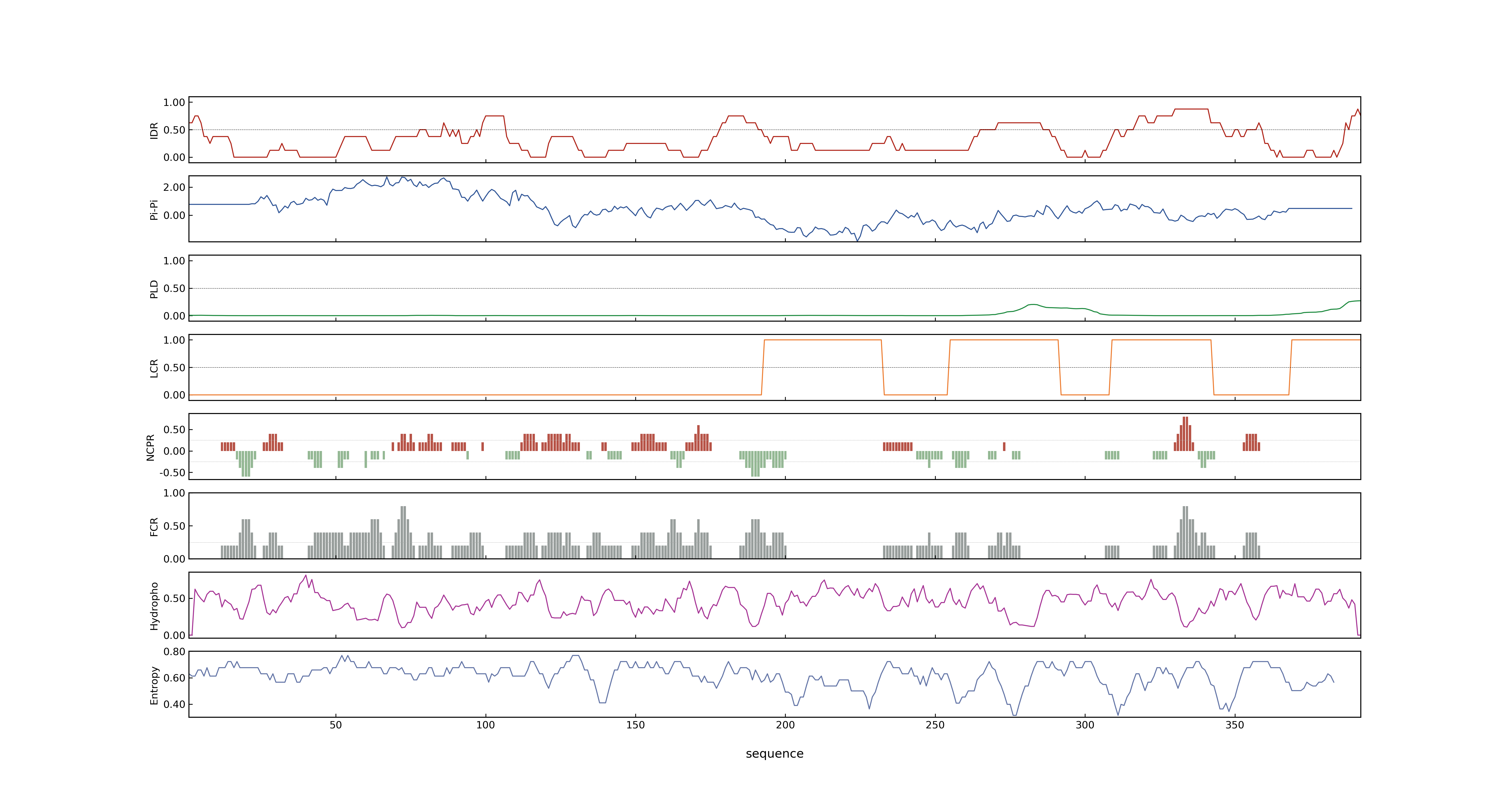
- PSP score
- LOC_Os03g21060.1: 0.8373
- PLAAC score
- LOC_Os03g21060.1: 0
- pLDDT score
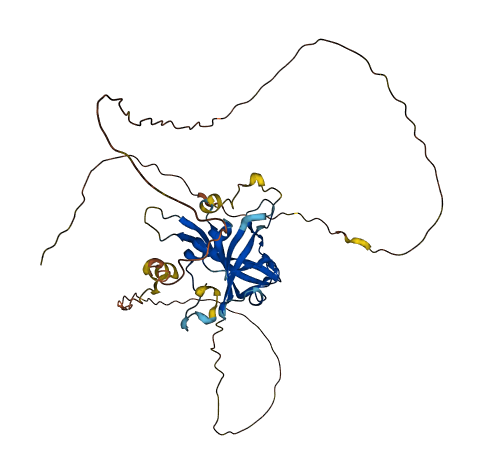
- 64.76
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os03g21060.1: 0.99982941
- MolPhase Result
- Publication
- The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway, 2014, Plant Cell Physiol.
- Identification and functional characterization of a rice NAC gene involved in the regulation of leaf senescence, 2013, BMC Plant Biol.
- OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice, 2014, Proceedings of the National Academy of Sciences.
-
Genbank accession number
- Key message
- OsNAP-overexpressing transgenic plants displayed an accelerated leaf senescence phenotype at the grain-filling stage, which might be caused by the elevated JA levels and the increased expression of the JA biosynthesis-related genes LOX2 and AOC1, and showed enhanced tolerance ability to MeJA treatment at the seedling stage
- Analysis of the OsNAP transcript levels in rice showed that this gene was significantly induced by ABA and abiotic stresses, including high salinity, drought and low temperature
- The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway
- RESULTS: In this study, we reported a member of the NAC transcription factor family named OsNAP whose expression is associated with leaf senescence, and investigated its preliminary function during the process of leaf senescence
- The results of qRT-PCR showed that the OsNAP transcripts were accumulated gradually in response to leaf senescence and treatment with methyl jasmonic acid (MeJA)
- Nevertheless, the leaf senescence process was delayed in OsNAP RNAi transgenic plants with a dramatic drop in JA levels and with decreased expression levels of the JA biosynthesis-related genes AOS2, AOC1 and OPR7
- CONCLUSIONS: These results suggest that OsNAP acts as a positive regulator of leaf senescence and this regulation may occur via the JA pathway
- Rice plants overexpressing OsNAP did not show growth retardation, but showed a significantly reduced rate of water loss, enhanced tolerance to high salinity, drought and low temperature at the vegetative stage, and improved yield under drought stress at the flowering stage
- Our data suggest that OsNAP functions as a transcriptional activator that plays a role in mediating abiotic stress responses in rice
- OsNAP is a member of the NAC transcription factor family; it is localized in the nucleus, and shows transcriptional activator activity in yeast
- Microarray analysis of transgenic plants overexpressing OsNAP revealed that many stress-related genes were up-regulated, including OsPP2C06/OsABI2, OsPP2C09, OsPP2C68 and OsSalT, and some genes coding for stress-related transcription factors (OsDREB1A, OsMYB2, OsAP37 and OsAP59)
- OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice
- Additionally, reduced OsNAP expression leads to delayed leaf senescence and an extended grain-filling period, resulting in a 6.3% and 10.3% increase in the grain yield of two independent representative RNAi lines, respectively.
- Thus, fine-tuning OsNAP expression should be a useful strategy for improving rice yield in the future
- Connection
- OsABI2~OsPP2C06~OsABIL1, OsNAP, The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway, Microarray analysis of transgenic plants overexpressing OsNAP revealed that many stress-related genes were up-regulated, including OsPP2C06/OsABI2, OsPP2C09, OsPP2C68 and OsSalT, and some genes coding for stress-related transcription factors (OsDREB1A, OsMYB2, OsAP37 and OsAP59)
- OsNAP, OsSIPP2C1~OsPP2C68~OsPP108, The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway, Microarray analysis of transgenic plants overexpressing OsNAP revealed that many stress-related genes were up-regulated, including OsPP2C06/OsABI2, OsPP2C09, OsPP2C68 and OsSalT, and some genes coding for stress-related transcription factors (OsDREB1A, OsMYB2, OsAP37 and OsAP59)
- OsNAP, OsPP2C09~OsPP15~OsPYL2, The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway, Microarray analysis of transgenic plants overexpressing OsNAP revealed that many stress-related genes were up-regulated, including OsPP2C06/OsABI2, OsPP2C09, OsPP2C68 and OsSalT, and some genes coding for stress-related transcription factors (OsDREB1A, OsMYB2, OsAP37 and OsAP59)
Prev Next