- Information
- Symbol: OsNMD3
- MSU: LOC_Os10g42320
- RAPdb: Os10g0573900
- PSP score
- LOC_Os10g42320.1: 0.1047
- PLAAC score
- LOC_Os10g42320.1: 0
- pLDDT score
- 83.66
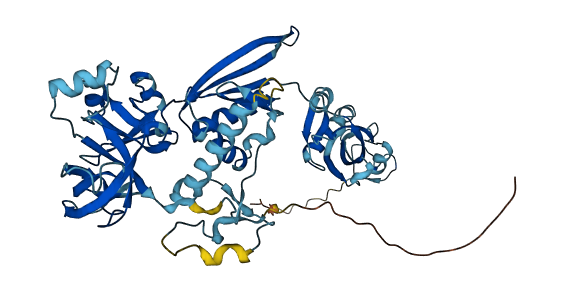
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os10g42320.1: 0.12960274
- MolPhase Result
- Publication
-
Genbank accession number
- Key message
- These expression profiles suggested that overexpression of OsNMD3(deltaNLS) affected ribosome biogenesis and certain basic pathways, leading to pleiotropic abnormalities in plant growth
- Analyses of the transactivation activity and cellulose biosynthesis level revealed low protein synthesis efficiency in the transgenic plants compared with the wild-type plants
- A dominant negative form of OsNMD3 with a truncated nuclear localization sequence (OsNMD3(deltaNLS)) was retained in the cytoplasm, consequently interfering with the release of OsNMD3 from pre-60S particles and disturbing the assembly of ribosome subunits
- Moreover, global expression profiles of the wild-type and transgenic plants were investigated using the Illumina RNA sequencing approach
- Retention of OsNMD3 in the cytoplasm disturbs protein synthesis efficiency and affects plant development in rice.
- sativa NMD3 (OsNMD3) shares all the common motifs and shuttles between the nucleus and cytoplasm via CRM1/XPO1
- This study chose a highly conserved trans-factor, the 60S ribosomal subunit nuclear export adaptor NMD3, to characterize the mechanism of ribosome biogenesis in the monocot plant Oryza sativa (rice)
- Connection
Prev Next