- Information
- Symbol: OsNSP1
- MSU: LOC_Os03g29480
- RAPdb: Os03g0408600
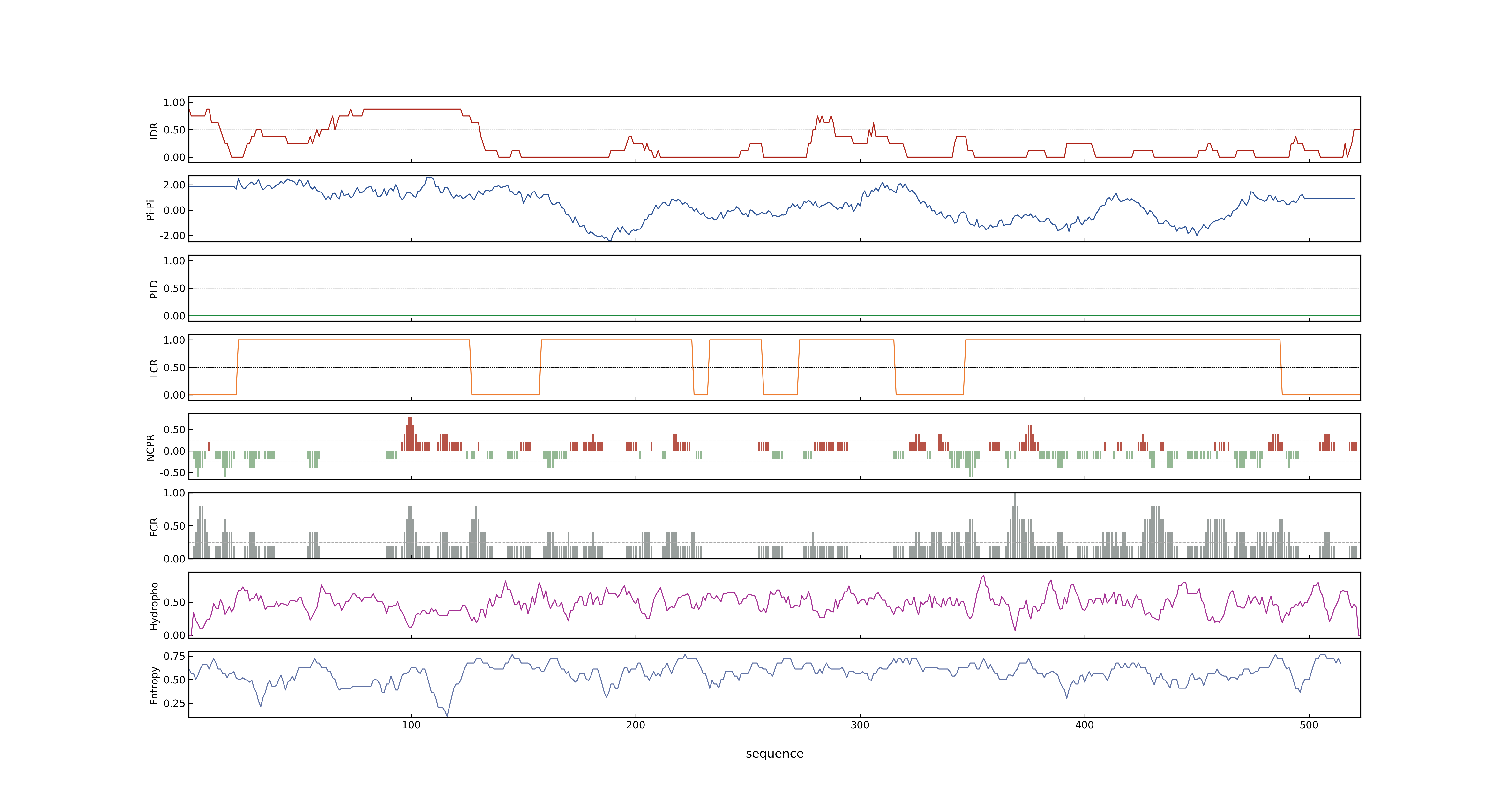
- PSP score
- LOC_Os03g29480.1: 0.5076
- PLAAC score
- LOC_Os03g29480.1: 0
- pLDDT score
- 75.74
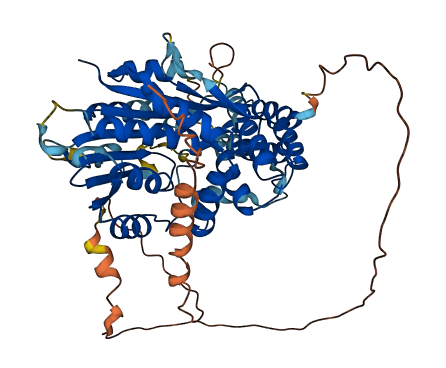
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os03g29480.1: 0.99873131
- MolPhase Result
- Publication
-
Genbank accession number
-
Key message
- Connection
- OsNSP1, OsNSP2, Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2, Legume GRAS (GAI, RGA, SCR)-type transcription factors NODULATION SIGNALING PATHWAY1 (NSP1) and NSP2 are essential for rhizobium Nod factor-induced nodulation
- OsNSP1, OsNSP2, Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2, However, legume NSP1 and NSP2 can be functionally replaced by nonlegume orthologs, including rice (Oryza sativa) NSP1 and NSP2, indicating that both proteins are functionally conserved in higher plants
- OsNSP1, OsNSP2, Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2, Here, we show that NSP1 and NSP2 are indispensable for strigolactone (SL) biosynthesis in the legume Medicago truncatula and in rice
- OsNSP1, OsNSP2, Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2, The disturbed SL biosynthesis in nsp1 nsp2 mutant backgrounds correlates with reduced expression of DWARF27, a gene essential for SL biosynthesis
- OsNSP1, OsNSP2, Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2, Therefore, we conclude that regulation of SL biosynthesis by NSP1 and NSP2 is an ancestral function conserved in higher plants
- OsNSP1, OsNSP2, Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2, NSP1 and NSP2 are single-copy genes in legumes, which implies that both proteins fulfill dual regulatory functions to control downstream targets after rhizobium-induced signaling as well as SL biosynthesis in nonsymbiotic conditions
- OsNSP1, OsNSP2, Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2, Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2
- OsNSP1, OsNSP2, Function of GRAS proteins in root nodule symbiosis is retained in homologs of a non-legume, rice, In this study, we demonstrated that their homologs, OsNSP1 and OsNSP2, from rice are able to fully rescue the RN symbiosis-defective phenotypes of the mutants of corresponding genes in the model legume, Lotus japonicus
- OsNSP1, OsNSP2, Function of GRAS proteins in root nodule symbiosis is retained in homologs of a non-legume, rice, Two GRAS-domain transcription factors, NSP1 and NSP2, have been shown to be required for RN symbiosis, but not for AM symbiosis
Prev Next