- Information
- Symbol: OsPHR2
- MSU: LOC_Os07g25710
- RAPdb: Os07g0438800
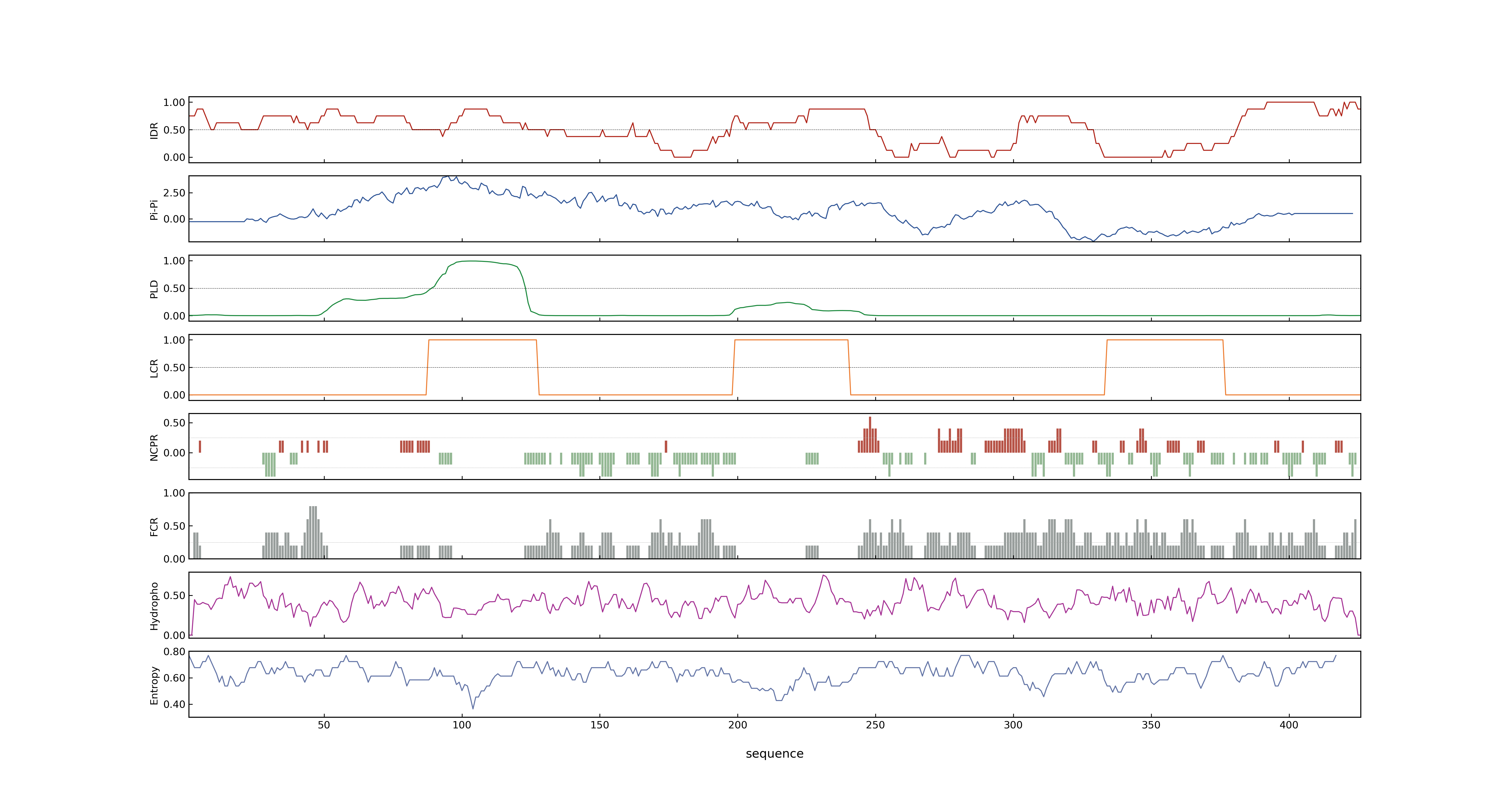
- PSP score
- LOC_Os07g25710.2: 0.9268
- LOC_Os07g25710.3: 0.9015
- LOC_Os07g25710.4: 0.5101
- LOC_Os07g25710.1: 0.9015
- PLAAC score
- LOC_Os07g25710.2: 0
- LOC_Os07g25710.3: 0
- LOC_Os07g25710.4: 0
- LOC_Os07g25710.1: 0
- pLDDT score
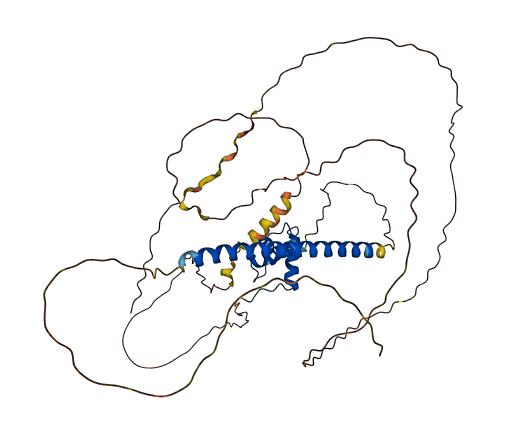
- 57.93
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os07g25710.1: 0.99994885
- LOC_Os07g25710.2: 0.99993228
- LOC_Os07g25710.3: 0.99994885
- LOC_Os07g25710.4: 0.99933546
- MolPhase Result
- Publication
- Molecular mechanisms regulating Pi-signaling and Pi homeostasis under OsPHR2, a central Pi-signaling regulator, in rice, 2011, Frontiers in Biology.
- Overexpression of OsPAP10a, a root-associated acid phosphatase, increased extracellular organic phosphorus utilization in rice, 2012, J Integr Plant Biol.
- OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice, 2010, Plant J.
- OsPHF1 regulates the plasma membrane localization of low- and high-affinity inorganic phosphate transporters and determines inorganic phosphate uptake and translocation in rice, 2011, Plant Physiol.
- Involvement of OsSPX1 in phosphate homeostasis in rice, 2009, Plant J.
- OsARF16, a transcription factor, is required for auxin and phosphate starvation response in rice Oryza sativa L., 2013, Plant Cell Environ.
- OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants, 2008, Plant Physiol.
- Investigating the contribution of the phosphate transport pathway to arsenic accumulation in rice, 2011, Plant Physiol.
- Role of OsPHR2 on phosphorus homeostasis and root hairs development in rice Oryza sativa L., 2008, Plant Signal Behav.
- Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner, 2014, Proc Natl Acad Sci U S A.
- SPX proteins regulate Pi homeostasis and signaling in different subcellular level., 2015, Plant Signal Behav.
- Genbank accession number
- Key message
- Under Pi-sufficient conditions, overexpression of OsPHR2 mimics Pi-starvation stress in rice with enhanced root elongation and proliferated root hair growth, suggesting the involvement of OsPHR2 in Pi-dependent root architecture alteration by both systematic and local pathways
- OsPT2 is responsible for most of the OsPHR2-mediated accumulation of excess Pi in shoots
- OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice
- Our results showed that both OsPHR1 and OsPHR2 are involved in Pi-starvation signaling pathway by regulation of the expression of Pi-starvation-induced genes, whereas only OsPHR2 overexpression results in the excessive accumulation of Pi in shoots under Pi-sufficient conditions
- OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants
- In hydroponic experiments, a rice mutant defective in OsPHF1 (for phosphate transporter traffic facilitator1) lost much of the ability to take up Pi and arsenate and to transport them from roots to shoots, whereas transgenic rice overexpressing either the Pi transporter OsPht1;8 (OsPT8) or the transcription factor OsPHR2 (for phosphate starvation response2) had enhanced abilities of Pi and arsenate uptake and translocation
- Mutation of OsPHF1 also reduced Pi accumulation in plants exhibiting excessive shoot Pi accumulation due to the overexpression of OsPHR2
- In OsPHR2-overexpression plants, some Pi transporters were up-regulated under Pi-sufficient conditions, which correlates with the strongly increased content of Pi
- The mechanism behind the OsPHR2 regulated Pi accumulation will provide useful approaches to develop smart plants with high Pi efficiency
- The results showed that both of the genes are involved in the Pi-signaling pathway, while overexpression of OsPHR2 mimics Pi-starvation stress with enhanced root elongation and proliferated root hairs, and results in the excessive accumulation of Pi in shoots under Pi-sufficient conditions
- OsPHR2 regulated proliferation of root hair growth and root elongation suggests that OsPHR2 is involved in both systematic and local Pi-signaling pathways
- Role of OsPHR2 on phosphorus homeostasis and root hairs development in rice (Oryza sativa L.)
- The transcript abundance of OsPAP10a is specifically induced by Pi deficiency and is controlled by OsPHR2, the central transcription factor controlling Pi homeostasis
- Here we review recent advances in our understanding of the OsPHR2-mediated phosphate-signaling pathway in rice
- OsSPX1 acts as a repressor in the OsPHR2-mediated phosphate-signaling pathway
- OsPHR2 positively regulates the low-affinity Pi transporter OsPT2 through physical interaction and reciprocal regulation of OsPHO2 in roots
- Some mutants screened from ethyl methanesulfonate (EMS)-mutagenized M2 population of OsPHR2 overexpression transgenic line removed the growth inhibition, indicating that some unknown factors are crucial for Pi utilization or plant growth under the regulation of OsPHR2
- Molecular mechanisms regulating Pi-signaling and Pi homeostasis under OsPHR2, a central Pi-signaling regulator, in rice
- Here we report that OsPHR2 positively regulates the low-affinity Pi transporter gene OsPT2 by physical interaction and upstream regulation of OsPHO2 in roots
- Our data also show that OsSPX1 is a negative regulator of OsPHR2 and is involved in the feedback of Pi-signaling network in roots that is defined by OsPHR2 and OsPHO2
- Previous reports have demonstrated that over-expression of OsPHR2 (the homolog of AtPHR1) and knockdown of OsSPX1 result in accumulation of excessive shoot Pi in rice
- OsSPX1 suppresses the regulation on expression of OsPT2 by OsPHR2 and the accumulation of excess shoot Pi, but it does not suppress induction of OsPT2 or the accumulation of excessive shoot Pi in the Ospho2 mutant
- Transgenic plants with overexpression and repression of OsPHR1 and OsPHR2, respectively, were used for investigation of roles of the genes in Pi-signaling pathway and Pi homeostasis under Pi-sufficient and deficient conditions
- OsPT2 is responsible for most of the OsPHR2-mediated accumulation of excess shoot Pi
- Suppression of OsSPX1 by RNA interference resulted in severe signs of toxicity caused by the over-accumulation of Pi, similar to that found in OsPHR2 (phosphate starvation response transcription factor 2) overexpressors and pho2 (phosphate-responsive mutant 2)
- Six phosphate starvation-induced genes (PSIs) were less induced by -Pi in osarf16 and these trends were similar to a knockdown mutant of OsPHR2 or AtPHR1, which was a key regulator under -Pi
- Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner
- AtPHR1 in Arabidopsis and OsPHR2 in rice are central regulators of Pi signaling, which regulates the expression of phosphate starvation-induced (PSI) genes by binding to the P1BS elements in the promoter of PSI genes
- Connection
- LTN1~OsPHO2, OsPHR2, Molecular mechanisms regulating Pi-signaling and Pi homeostasis under OsPHR2, a central Pi-signaling regulator, in rice, OsPHR2 positively regulates the low-affinity Pi transporter OsPT2 through physical interaction and reciprocal regulation of OsPHO2 in roots
- OsPHR2, OsPht1;2~OsPT2, Molecular mechanisms regulating Pi-signaling and Pi homeostasis under OsPHR2, a central Pi-signaling regulator, in rice, OsPHR2 positively regulates the low-affinity Pi transporter OsPT2 through physical interaction and reciprocal regulation of OsPHO2 in roots
- OsPHR2, OsPht1;2~OsPT2, Molecular mechanisms regulating Pi-signaling and Pi homeostasis under OsPHR2, a central Pi-signaling regulator, in rice, OsPT2 is responsible for most of the OsPHR2-mediated accumulation of excess Pi in shoots
- OsPHR2, OsSPX1, Molecular mechanisms regulating Pi-signaling and Pi homeostasis under OsPHR2, a central Pi-signaling regulator, in rice, OsSPX1 acts as a repressor in the OsPHR2-mediated phosphate-signaling pathway
- OsPAP10a, OsPHR2, Overexpression of OsPAP10a, a root-associated acid phosphatase, increased extracellular organic phosphorus utilization in rice, The transcript abundance of OsPAP10a is specifically induced by Pi deficiency and is controlled by OsPHR2, the central transcription factor controlling Pi homeostasis
- LTN1~OsPHO2, OsPHR2, OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice, Here we report that OsPHR2 positively regulates the low-affinity Pi transporter gene OsPT2 by physical interaction and upstream regulation of OsPHO2 in roots
- LTN1~OsPHO2, OsPHR2, OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice, OsSPX1 suppresses the regulation on expression of OsPT2 by OsPHR2 and the accumulation of excess shoot Pi, but it does not suppress induction of OsPT2 or the accumulation of excessive shoot Pi in the Ospho2 mutant
- LTN1~OsPHO2, OsPHR2, OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice, Our data also show that OsSPX1 is a negative regulator of OsPHR2 and is involved in the feedback of Pi-signaling network in roots that is defined by OsPHR2 and OsPHO2
- OsPHR2, OsSPX1, OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice, Previous reports have demonstrated that over-expression of OsPHR2 (the homolog of AtPHR1) and knockdown of OsSPX1 result in accumulation of excessive shoot Pi in rice
- OsPHR2, OsSPX1, OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice, OsSPX1 suppresses the regulation on expression of OsPT2 by OsPHR2 and the accumulation of excess shoot Pi, but it does not suppress induction of OsPT2 or the accumulation of excessive shoot Pi in the Ospho2 mutant
- OsPHR2, OsSPX1, OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice, Our data also show that OsSPX1 is a negative regulator of OsPHR2 and is involved in the feedback of Pi-signaling network in roots that is defined by OsPHR2 and OsPHO2
- OsPHR2, OsSPX1, OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice, OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice
- OsPHR2, OsPht1;2~OsPT2, OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice, Here we report that OsPHR2 positively regulates the low-affinity Pi transporter gene OsPT2 by physical interaction and upstream regulation of OsPHO2 in roots
- OsPHR2, OsPht1;2~OsPT2, OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice, OsPT2 is responsible for most of the OsPHR2-mediated accumulation of excess shoot Pi
- OsPHR2, OsPht1;2~OsPT2, OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice, OsSPX1 suppresses the regulation on expression of OsPT2 by OsPHR2 and the accumulation of excess shoot Pi, but it does not suppress induction of OsPT2 or the accumulation of excessive shoot Pi in the Ospho2 mutant
- OsPHR2, OsPht1;2~OsPT2, OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice, OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice
- OsPHF1, OsPHR2, OsPHF1 regulates the plasma membrane localization of low- and high-affinity inorganic phosphate transporters and determines inorganic phosphate uptake and translocation in rice, Mutation of OsPHF1 also reduced Pi accumulation in plants exhibiting excessive shoot Pi accumulation due to the overexpression of OsPHR2
- OsPHF1, OsPHR2, OsPHF1 regulates the plasma membrane localization of low- and high-affinity inorganic phosphate transporters and determines inorganic phosphate uptake and translocation in rice, However, the transcript level of OsPHF1 itself is not controlled by OsPHR2
- LTN1~OsPHO2, OsPHR2, Involvement of OsSPX1 in phosphate homeostasis in rice, Suppression of OsSPX1 by RNA interference resulted in severe signs of toxicity caused by the over-accumulation of Pi, similar to that found in OsPHR2 (phosphate starvation response transcription factor 2) overexpressors and pho2 (phosphate-responsive mutant 2)
- LTN1~OsPHO2, OsPHR2, Involvement of OsSPX1 in phosphate homeostasis in rice, Quantitative RT-PCR showed that expression of OsSPX1 was strongly induced in OsPHR2 overexpression and pho2 mutant plants, indicating that OsSPX1 occurs downstream of OsPHR2 and PHO2
- OsPHR2, OsSPX1, Involvement of OsSPX1 in phosphate homeostasis in rice, Suppression of OsSPX1 by RNA interference resulted in severe signs of toxicity caused by the over-accumulation of Pi, similar to that found in OsPHR2 (phosphate starvation response transcription factor 2) overexpressors and pho2 (phosphate-responsive mutant 2)
- OsPHR2, OsSPX1, Involvement of OsSPX1 in phosphate homeostasis in rice, Quantitative RT-PCR showed that expression of OsSPX1 was strongly induced in OsPHR2 overexpression and pho2 mutant plants, indicating that OsSPX1 occurs downstream of OsPHR2 and PHO2
- OsARF16, OsPHR2, OsARF16, a transcription factor, is required for auxin and phosphate starvation response in rice Oryza sativa L., Six phosphate starvation-induced genes (PSIs) were less induced by -Pi in osarf16 and these trends were similar to a knockdown mutant of OsPHR2 or AtPHR1, which was a key regulator under -Pi
- OsPHR1~PHR1, OsPHR2, OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants, In this work, two OsPHR genes from rice (Oryza sativa) were isolated and designated as OsPHR1 and OsPHR2 based on amino acid sequence homology to AtPHR1
- OsPHR1~PHR1, OsPHR2, OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants, Our results showed that both OsPHR1 and OsPHR2 are involved in Pi-starvation signaling pathway by regulation of the expression of Pi-starvation-induced genes, whereas only OsPHR2 overexpression results in the excessive accumulation of Pi in shoots under Pi-sufficient conditions
- OsPHF1, OsPHR2, Investigating the contribution of the phosphate transport pathway to arsenic accumulation in rice, In hydroponic experiments, a rice mutant defective in OsPHF1 (for phosphate transporter traffic facilitator1) lost much of the ability to take up Pi and arsenate and to transport them from roots to shoots, whereas transgenic rice overexpressing either the Pi transporter OsPht1;8 (OsPT8) or the transcription factor OsPHR2 (for phosphate starvation response2) had enhanced abilities of Pi and arsenate uptake and translocation
- OsPHR2, OsPht1;8~OsPT8, Investigating the contribution of the phosphate transport pathway to arsenic accumulation in rice, In hydroponic experiments, a rice mutant defective in OsPHF1 (for phosphate transporter traffic facilitator1) lost much of the ability to take up Pi and arsenate and to transport them from roots to shoots, whereas transgenic rice overexpressing either the Pi transporter OsPht1;8 (OsPT8) or the transcription factor OsPHR2 (for phosphate starvation response2) had enhanced abilities of Pi and arsenate uptake and translocation
- OsPHR1~PHR1, OsPHR2, Role of OsPHR2 on phosphorus homeostasis and root hairs development in rice Oryza sativa L., Two PHR genes were isolated from rice and designated as OsPHR1 and OsPHR2 based on amino acid sequence homology to AtPHR1
- OsPHR1~PHR1, OsPHR2, Role of OsPHR2 on phosphorus homeostasis and root hairs development in rice Oryza sativa L., Transgenic plants with overexpression and repression of OsPHR1 and OsPHR2, respectively, were used for investigation of roles of the genes in Pi-signaling pathway and Pi homeostasis under Pi-sufficient and deficient conditions
- OsPHR2, OsSPX4, SPX4 Negatively Regulates Phosphate Signaling and Homeostasis through Its Interaction with PHR2 in Rice, SPX4 Negatively Regulates Phosphate Signaling and Homeostasis through Its Interaction with PHR2 in Rice
- OsPHR2, OsSPX1, Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner, Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner
- OsPHR2, OsSPX2, Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner, Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner
- OsPHR2, OsSPX2, Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner, Indeed, SPX1 and SPX2 proteins interact with PHR2 through their SPX domain, inhibiting its binding to P1BS (the PHR1-binding sequence: GNATATNC)
- OsPHR2, OsSPX1, Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner, Indeed, SPX1 and SPX2 proteins interact with PHR2 through their SPX domain, inhibiting its binding to P1BS (the PHR1-binding sequence: GNATATNC)
- OsPHR1~PHR1, OsPHR2, Genetic manipulation of a high-affinity PHR1 target cis-element to improve phosphorous uptake in Oryza sativa L., In this work, we investigated the DNA binding affinity of the PHR1 ortholog OsPHR2 to its downstream target genes in Oryza sativa (rice)
- OsPHR2, OsPHR3, Integrative comparison of the role of the PHR1 subfamily in phosphate signaling and homeostasis in rice., Here, we identified functional redundancy among three PHR1 orthologs in rice, OsPHR1, OsPHR2 and OsPHR3, using phylogenetic and mutation analysis
- OsPHR2, OsPHR3, Integrative comparison of the role of the PHR1 subfamily in phosphate signaling and homeostasis in rice., OsPHR3, in conjunction with OsPHR1 and OsPHR2, function in transcriptional activation of most Pi starvation-induced (PSI) genes
- OsPHR2, OsPHR3, Integrative comparison of the role of the PHR1 subfamily in phosphate signaling and homeostasis in rice., We propose that OsPHR1, OsPHR2 and OsPHR3 form a network and play diverse roles in regulating Pi signaling and homeostasis in rice
- OsPHR2, OsSPX1, Down-regulation of OsSPX1 caused semi-male sterility, resulting in reduction of grain yield in rice., OsSPX1, a rice SPX domain gene, involved in the phosphate (Pi)-sensing mechanism plays an essential role in the Pi-signalling network through interaction with OsPHR2
- OsLPR3, OsPHR2, Identification and expression analysis of OsLPR family revealed the potential roles of OsLPR3 and 5 in maintaining phosphate homeostasis in rice., Strong induction in the relative expression levels of OsLPR3 and 5 in osphr2 suggested their negative transcriptional regulation by OsPHR2
- OsPHR2, OsPHR4, Phosphate starvation induced OsPHR4 mediates Pi-signaling and homeostasis in rice., The expression of OsPHR4 is positively regulated by OsPHR1, OsPHR2 and OsPHR3
- OsPAP21b, OsPHR2, Improvement of Phosphate Acquisition and Utilization by a Secretory Purple Acid Phosphatase OsPAP21b in Rice., OsPAP21b was found to be under the transcriptional control of OsPHR2 and strictly regulated by plant Pi-status at both transcript and protein levels
- OsNLA1, OsPHR2, OsNLA1, a RING-type ubiquitin ligase, maintains phosphate homeostasis in Oryza sativa via degradation of phosphate transporters., In contrast to what has been observed in Arabidopsis, the transcript abundance of OsNLA1 did not decrease under Pi limited conditions or in OsmiR827 (microRNA827)- or OsPHR2 (PHOSPHATE STARVATION RESPONSE 2)-overexpressing transgenic lines
Prev Next