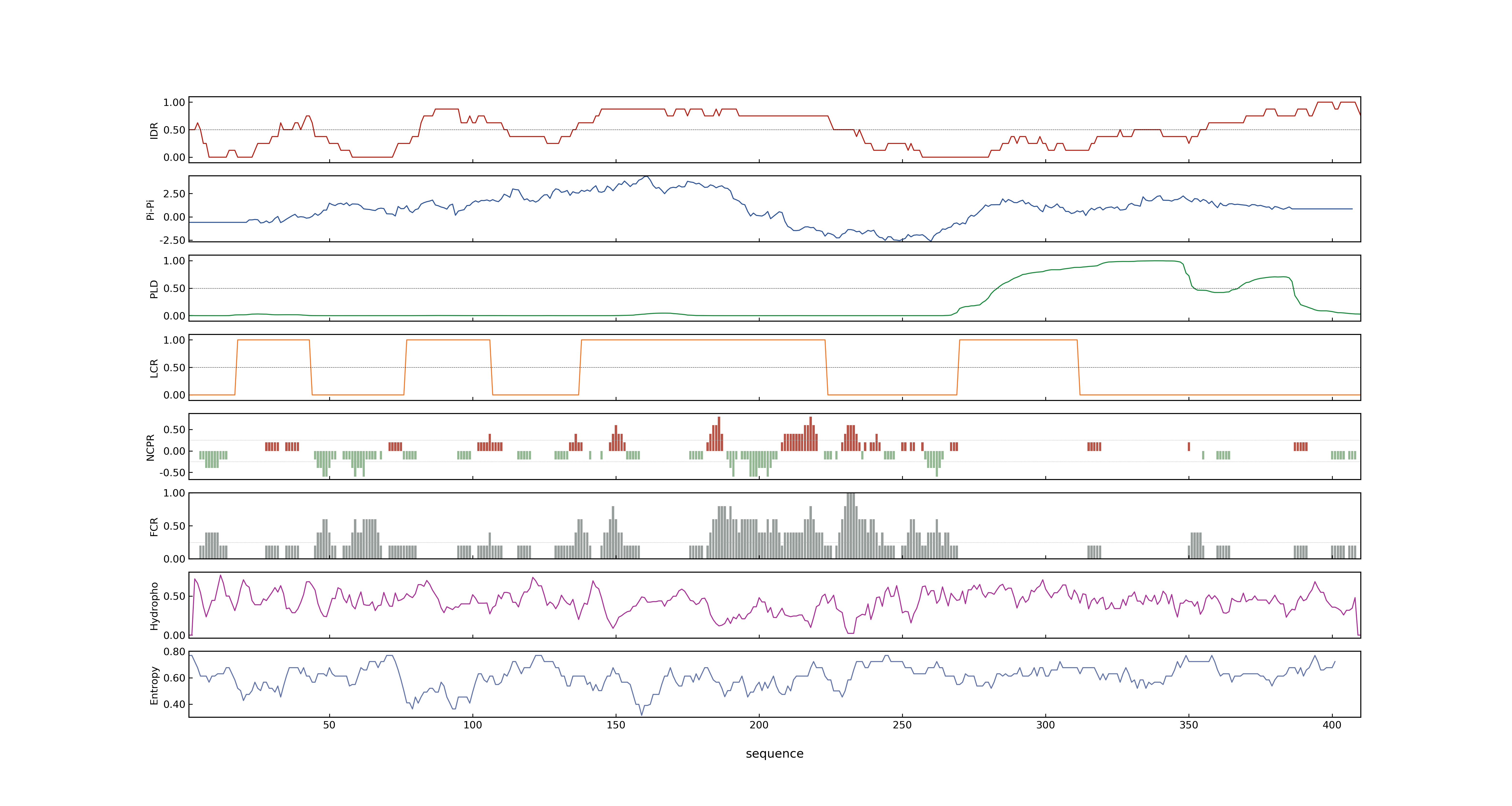
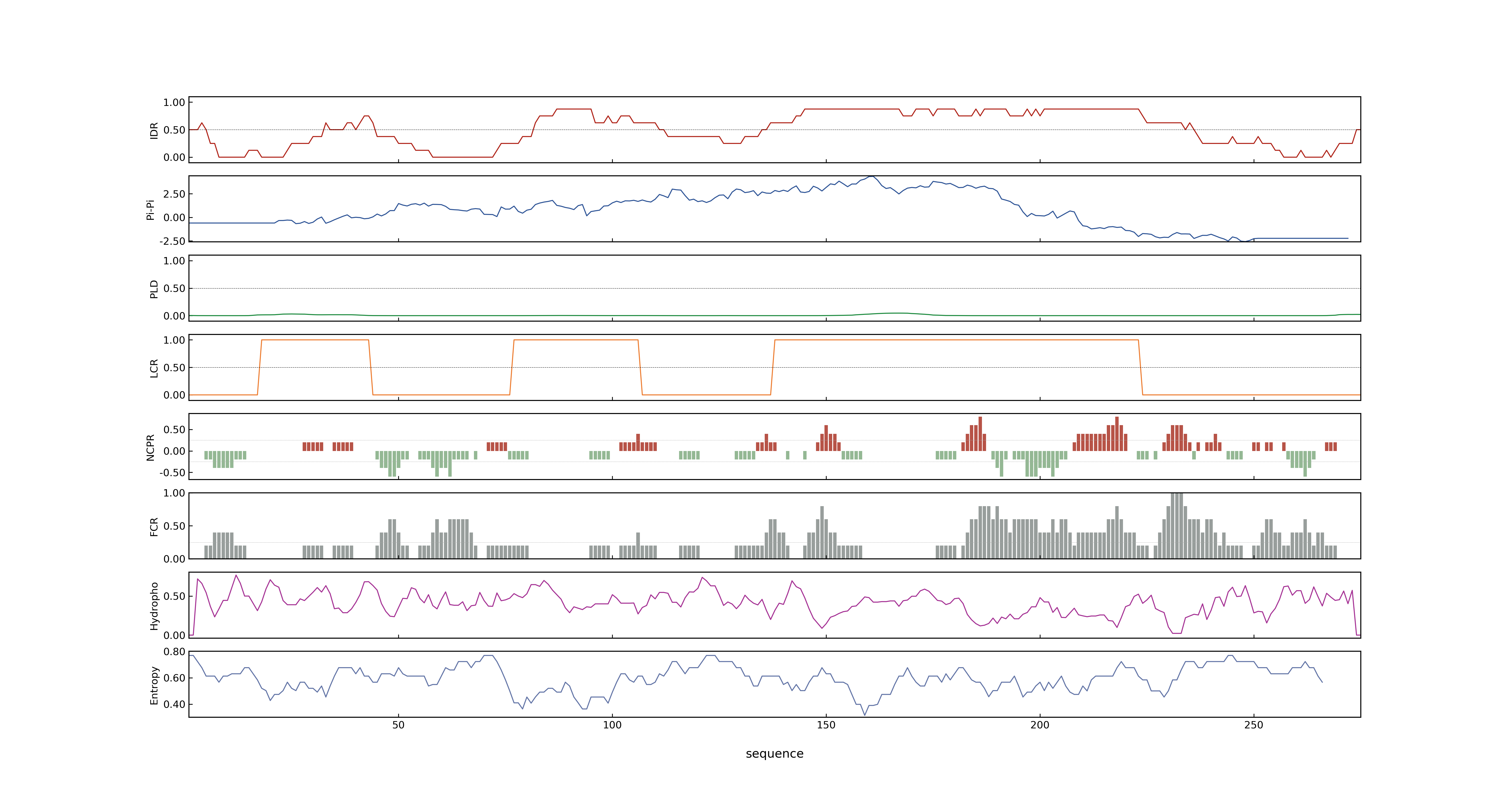
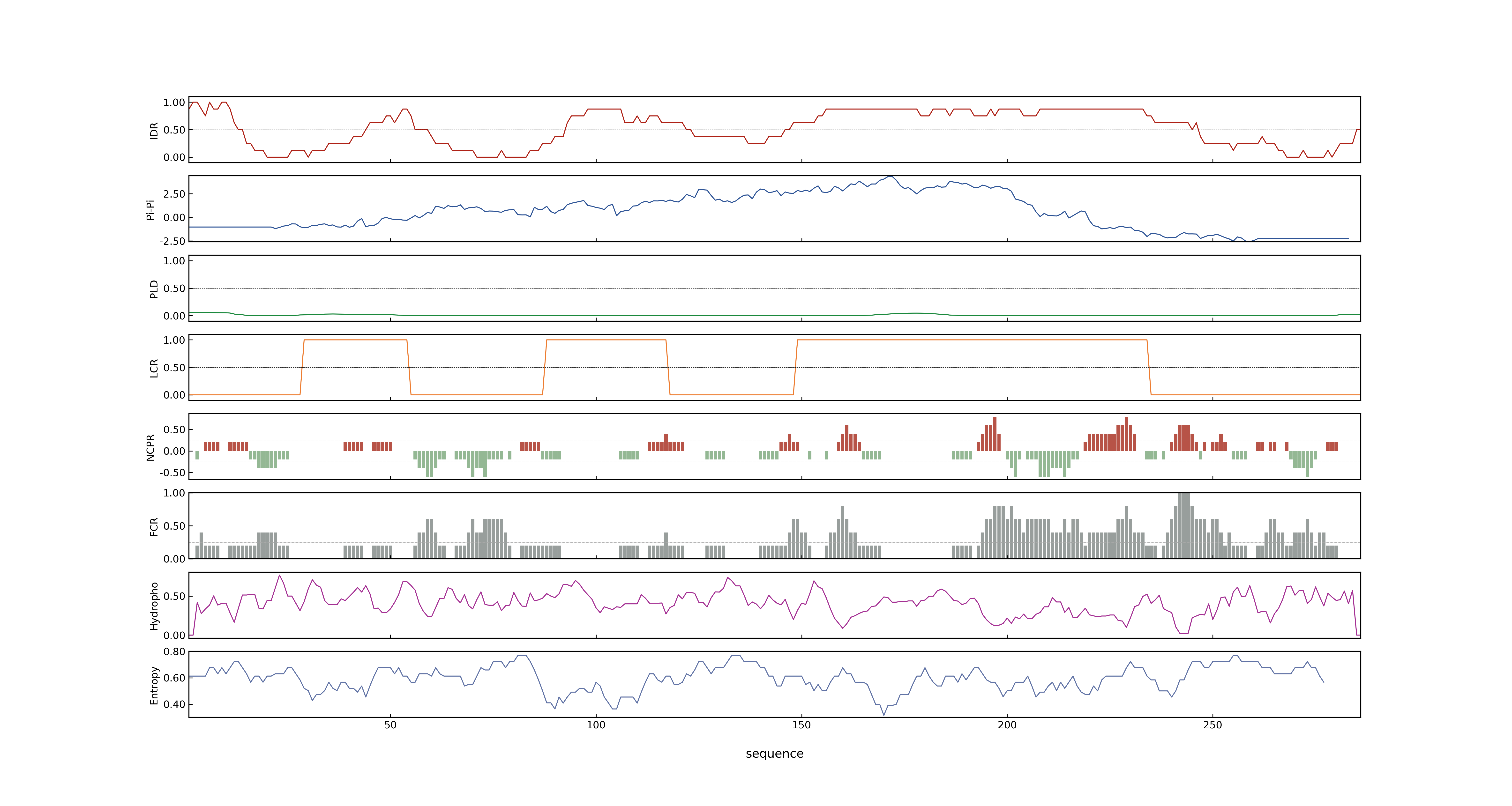
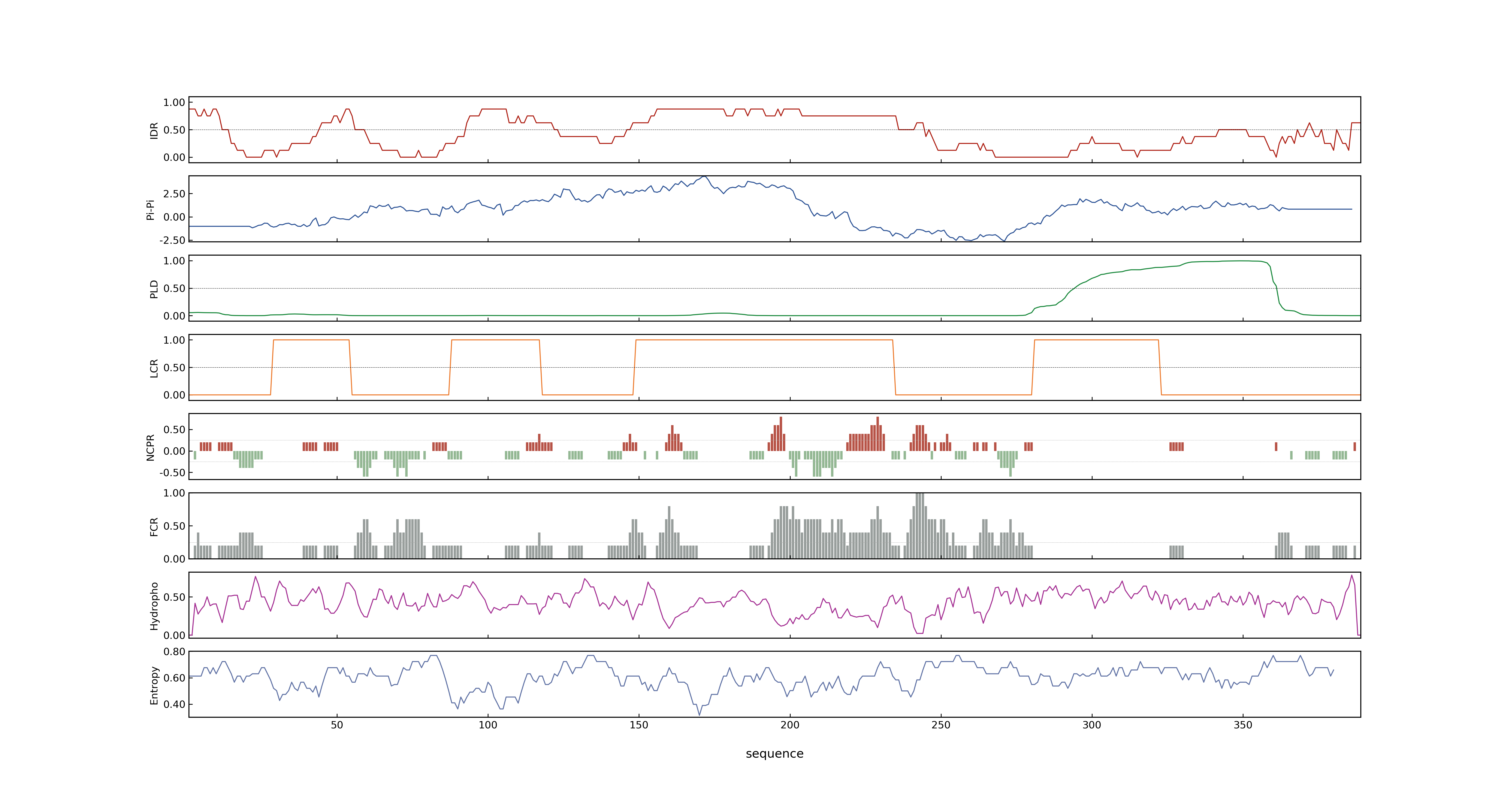
- Information
- Symbol: OsPIL13,OsPIL1
- MSU: LOC_Os03g56950
- RAPdb: Os03g0782500
- PSP score
- LOC_Os03g56950.7: 0.9082
- LOC_Os03g56950.5: 0.6671
- LOC_Os03g56950.4: 0.9028
- LOC_Os03g56950.1: 0.9305
- LOC_Os03g56950.6: 0.922
- LOC_Os03g56950.2: 0.9305
- LOC_Os03g56950.3: 0.9028
- PLAAC score
- LOC_Os03g56950.7: 14.512
- LOC_Os03g56950.5: 0
- LOC_Os03g56950.4: 15.709
- LOC_Os03g56950.1: 15.709
- LOC_Os03g56950.6: 0
- LOC_Os03g56950.2: 15.709
- LOC_Os03g56950.3: 15.709
(Zhao et al., New Biotechnol, 2011, 28:788-797)
- pLDDT score
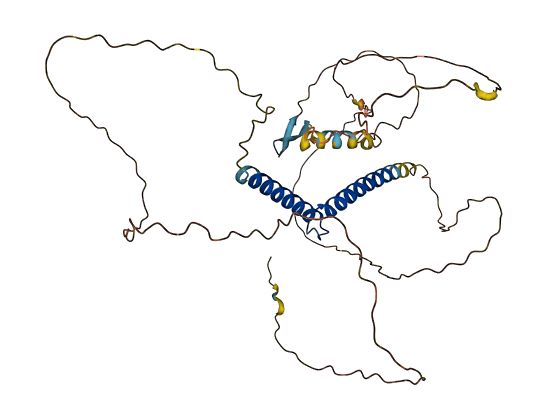
- 57.42
- Protein Structure from AlphaFold and UniProt
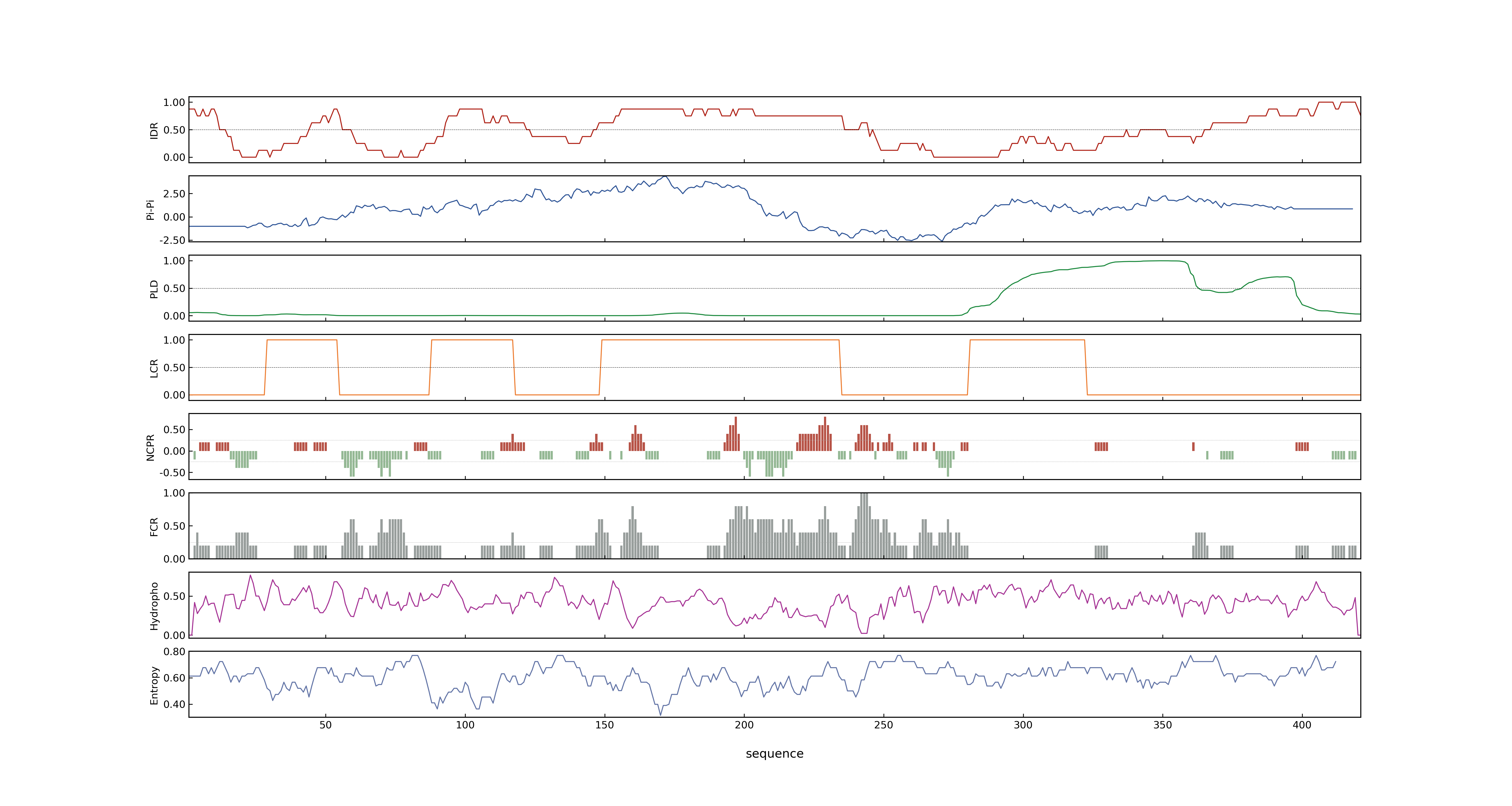
- MolPhase score
- LOC_Os03g56950.1: 0.99981743
- LOC_Os03g56950.2: 0.99981743
- LOC_Os03g56950.3: 0.99983084
- LOC_Os03g56950.4: 0.99983084
- LOC_Os03g56950.5: 0.99866890
- LOC_Os03g56950.6: 0.99905425
- LOC_Os03g56950.7: 0.99979120
- MolPhase Result
- Publication
- An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, 2011, New Biotechnol.
- Characterization of a Set of Phytochrome-Interacting Factor-Like bHLH Proteins inOryza sativa, 2014, Bioscience, Biotechnology and Biochemistry.
- Rice phytochrome-interacting factor-like protein OsPIL1 functions as a key regulator of internode elongation and induces a morphological response to drought stress, 2012, Proc Natl Acad Sci U S A.
- Double overexpression of DREB and PIF transcription factors improves drought stress tolerance and cell elongation in transgenic plants., 2016, Plant Biotechnol J.
- Roles of rice PHYTOCHROME-INTERACTING FACTOR-LIKE1 OsPIL1 in leaf senescence., 2017, Plant Signal Behav.
- Rice Phytochrome-Interacting Factor-Like1 OsPIL1 is involved in the promotion of chlorophyll biosynthesis through feed-forward regulatory loops., 2017, J Exp Bot.
- Genbank accession number
- Key message
- Oligoarray analysis revealed OsPIL1 downstream genes, which were enriched for cell wall-related genes responsible for cell elongation
- These data suggest that OsPIL1 functions as a key regulatory factor of reduced plant height via cell wall-related genes in response to drought stress
- We found that OsPIL1 was highly expressed in the node portions of the stem using promoter-glucuronidase analysis
- In addition, OsPIL13 and OsPIL15 colocalize with OsPRR1, an ortholog of the Arabidopsis APRR1 gene that controls photoperiodic flowering response through clock function
- Together, these results suggest that overexpression of OsLF might repress expression of OsGI and Hd1 by competing with OsPRR1 in interacting with OsPIL13 and OsPIL15 and thus induce late flowering
- An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15
- Here, we demonstrate that a phytochrome-interacting factor-like protein, OsPIL1/OsPIL13, acts as a key regulator of reduced internode elongation in rice under drought conditions
- Under drought stress conditions, OsPIL1 expression was inhibited during the light period
- Rice phytochrome-interacting factor-like protein OsPIL1 functions as a key regulator of internode elongation and induces a morphological response to drought stress
- It was found that the expression of OsPIL13 is under the control of circadian rhythms (clock), while the expression of OsPIL15 is negatively regulated by light upon the onset to light exposure of etiolated seedlings
- The level of OsPIL1 mRNA in rice seedlings grown under nonstressed conditions with light/dark cycles oscillated in a circadian manner with peaks in the middle of the light period
- Transcriptome analyses showed increased expression of abiotic stress-inducible DREB1A downstream genes and cell elongation-related OsPIL1 downstream genes in the double overexpressors, which suggests that these two transcription factors function independently in the transgenic plants despite the trade-offs required to balance plant growth and stress tolerance
- The transgenic plants overexpressing both OsPIL1 and DREB1A showed improved drought stress tolerance similar to that of DREB1A overexpressors
- OsPIL1 is a rice homologue of Arabidopsis PHYTOCHROME-INTERACTING FACTOR 4 (PIF4), and it enhances cell elongation by activating cell wall-related gene expression
- Here, we show that OsPIL1 negatively regulates leaf senescence in rice
- Microarray analysis revealed that during DIS, a number of senescence-associated genes were upregulated and OsGLKs, negative regulators of leaf senescence, were strongly repressed in ospil1 mutants
- In addition, OsPIL1 expressed in Arabidopsis upregulates the transcription of ORESARA1, a major senescence-inducing NAC transcription factor and one of the downstream genes of Arabidopsis PIF4, by directly binding the promoter region
- During dark-induced senescence (DIS), ospil1 mutants senesced earlier than wild type; this is opposite to mutants of Arabidopsis PIF4 and PIF5, the closest homologs of OsPIL1
- In addition, OsPIL1 directly up-regulates the expression of two transcription factor genes, GOLDEN2-LIKE1 (OsGLK1) and OsGLK2
- Transcriptome analysis revealed that several genes responsible for Chl biosynthesis and photosynthesis were significantly down-regulated in ospil1 leaves
- Thus, OsPIL1 is involved in the promotion of Chl biosynthesis by up-regulating the transcription of OsPORB and OsCAO1 via trifurcate feed-forward regulatory loops involving two OsGLKs
- Connection
- OsGI, OsPIL13~OsPIL1, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Together, these results suggest that overexpression of OsLF might repress expression of OsGI and Hd1 by competing with OsPRR1 in interacting with OsPIL13 and OsPIL15 and thus induce late flowering
- OsPIL13~OsPIL1, OsPRR1, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, In addition, OsPIL13 and OsPIL15 colocalize with OsPRR1, an ortholog of the Arabidopsis APRR1 gene that controls photoperiodic flowering response through clock function
- OsPIL13~OsPIL1, OsPRR1, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Together, these results suggest that overexpression of OsLF might repress expression of OsGI and Hd1 by competing with OsPRR1 in interacting with OsPIL13 and OsPIL15 and thus induce late flowering
- Hd1, OsPIL13~OsPIL1, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Together, these results suggest that overexpression of OsLF might repress expression of OsGI and Hd1 by competing with OsPRR1 in interacting with OsPIL13 and OsPIL15 and thus induce late flowering
- OsLF, OsPIL13~OsPIL1, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Yeast two hybrid and colocalization assays revealed that OsLF interacts strongly with OsPIL13 and OsPIL15 that are potentially involved in light signaling
- OsLF, OsPIL13~OsPIL1, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Together, these results suggest that overexpression of OsLF might repress expression of OsGI and Hd1 by competing with OsPRR1 in interacting with OsPIL13 and OsPIL15 and thus induce late flowering
- OsLF, OsPIL13~OsPIL1, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15
- OsPIL13~OsPIL1, OsPIL15, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Yeast two hybrid and colocalization assays revealed that OsLF interacts strongly with OsPIL13 and OsPIL15 that are potentially involved in light signaling
- OsPIL13~OsPIL1, OsPIL15, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, In addition, OsPIL13 and OsPIL15 colocalize with OsPRR1, an ortholog of the Arabidopsis APRR1 gene that controls photoperiodic flowering response through clock function
- OsPIL13~OsPIL1, OsPIL15, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, Together, these results suggest that overexpression of OsLF might repress expression of OsGI and Hd1 by competing with OsPRR1 in interacting with OsPIL13 and OsPIL15 and thus induce late flowering
- OsPIL13~OsPIL1, OsPIL15, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15, An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15
- OsPIL13~OsPIL1, OsPRR1, Characterization of a Set of Phytochrome-Interacting Factor-Like bHLH Proteins inOryza sativa, Some of them (OsPIL11, OsPIL12, and OsPIL13) showed the ability to interact with the putative OsPRR1 (PSEUDO-RESPONSE REGULATOR 1) clock component, as far as the results of yeast two-hybrid assays were concerned
- OsPIL13~OsPIL1, OsPIL15, Characterization of a Set of Phytochrome-Interacting Factor-Like bHLH Proteins inOryza sativa, It was found that the expression of OsPIL13 is under the control of circadian rhythms (clock), while the expression of OsPIL15 is negatively regulated by light upon the onset to light exposure of etiolated seedlings
- OsPIL13~OsPIL1, OsPIL15, Characterization of a Set of Phytochrome-Interacting Factor-Like bHLH Proteins inOryza sativa, When the rice genes (OsPIL11 to OsPIL15) were over-expressed in A
- OsPIL12, OsPIL13~OsPIL1, Characterization of a Set of Phytochrome-Interacting Factor-Like bHLH Proteins inOryza sativa, Some of them (OsPIL11, OsPIL12, and OsPIL13) showed the ability to interact with the putative OsPRR1 (PSEUDO-RESPONSE REGULATOR 1) clock component, as far as the results of yeast two-hybrid assays were concerned
- OsPIL11, OsPIL13~OsPIL1, Characterization of a Set of Phytochrome-Interacting Factor-Like bHLH Proteins inOryza sativa, In this study, we identified and characterized a set of highly homologous members (designated OsPIL11 to OsPIL16) in the model monocotyledon rice (Oryza sativa)
- OsPIL11, OsPIL13~OsPIL1, Characterization of a Set of Phytochrome-Interacting Factor-Like bHLH Proteins inOryza sativa, Some of them (OsPIL11, OsPIL12, and OsPIL13) showed the ability to interact with the putative OsPRR1 (PSEUDO-RESPONSE REGULATOR 1) clock component, as far as the results of yeast two-hybrid assays were concerned
- OsPIL11, OsPIL13~OsPIL1, Characterization of a Set of Phytochrome-Interacting Factor-Like bHLH Proteins inOryza sativa, When the rice genes (OsPIL11 to OsPIL15) were over-expressed in A
- OsCAO1~PGL, OsPIL13~OsPIL1, Rice Phytochrome-Interacting Factor-Like1 OsPIL1 is involved in the promotion of chlorophyll biosynthesis through feed-forward regulatory loops., Using promoter binding and transactivation assays, we found that OsPIL1 binds to the promoters of two Chl biosynthetic genes, OsPORB and OsCAO1, and promotes their transcription
- OsCAO1~PGL, OsPIL13~OsPIL1, Rice Phytochrome-Interacting Factor-Like1 OsPIL1 is involved in the promotion of chlorophyll biosynthesis through feed-forward regulatory loops., Thus, OsPIL1 is involved in the promotion of Chl biosynthesis by up-regulating the transcription of OsPORB and OsCAO1 via trifurcate feed-forward regulatory loops involving two OsGLKs
- OsGLK1, OsPIL13~OsPIL1, Rice Phytochrome-Interacting Factor-Like1 OsPIL1 is involved in the promotion of chlorophyll biosynthesis through feed-forward regulatory loops., In addition, OsPIL1 directly up-regulates the expression of two transcription factor genes, GOLDEN2-LIKE1 (OsGLK1) and OsGLK2
- OsPIL13~OsPIL1, OsPORB~FGL, Rice Phytochrome-Interacting Factor-Like1 OsPIL1 is involved in the promotion of chlorophyll biosynthesis through feed-forward regulatory loops., Using promoter binding and transactivation assays, we found that OsPIL1 binds to the promoters of two Chl biosynthetic genes, OsPORB and OsCAO1, and promotes their transcription
- OsPIL13~OsPIL1, OsPORB~FGL, Rice Phytochrome-Interacting Factor-Like1 OsPIL1 is involved in the promotion of chlorophyll biosynthesis through feed-forward regulatory loops., Thus, OsPIL1 is involved in the promotion of Chl biosynthesis by up-regulating the transcription of OsPORB and OsCAO1 via trifurcate feed-forward regulatory loops involving two OsGLKs