- Information
- Symbol: OsPORA
- MSU: LOC_Os04g58200
- RAPdb: Os04g0678700
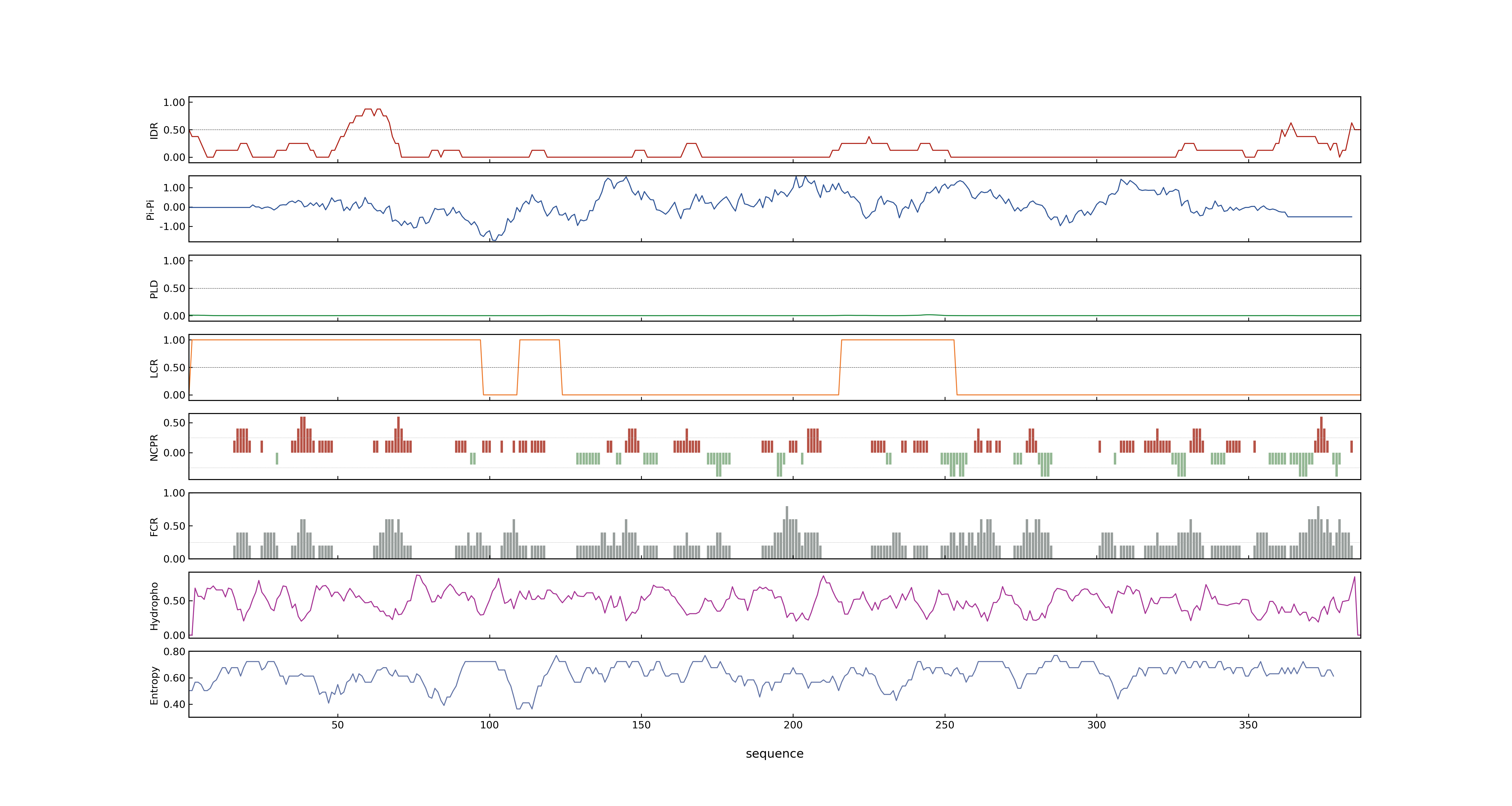
- PSP score
- LOC_Os04g58200.1: 0.0699
- LOC_Os04g58200.3: 0.0444
- LOC_Os04g58200.2: 0.1304
- PLAAC score
- LOC_Os04g58200.1: 0
- LOC_Os04g58200.3: 0
- LOC_Os04g58200.2: 0
- pLDDT score
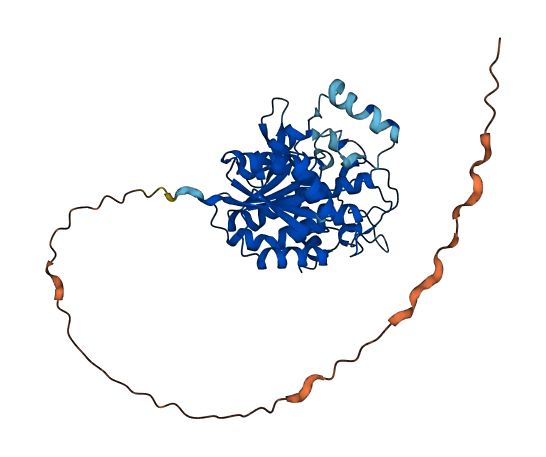
- 84.23
- Protein Structure from AlphaFold and UniProt
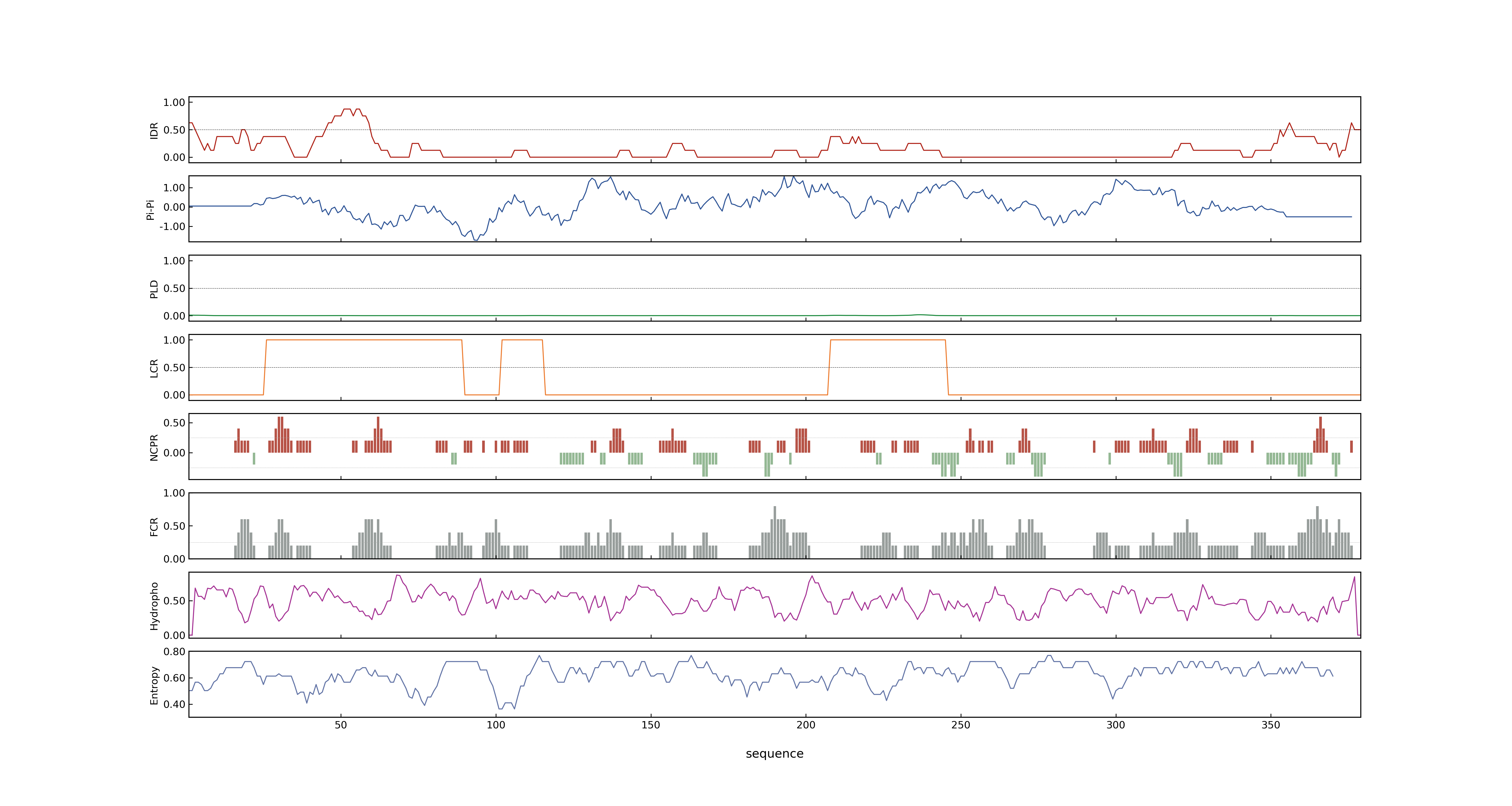
- MolPhase score
- LOC_Os04g58200.1: 0.13308593
- LOC_Os04g58200.2: 0.11522512
- LOC_Os04g58200.3: 0.00368741
- MolPhase Result
- Publication
- NOA1 functions in a temperature-dependent manner to regulate chlorophyll biosynthesis and Rubisco formation in rice, 2011, PLoS One.
- The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions, 2013, Plant J.
- Two NADPH: Protochlorophyllide Oxidoreductase POR Isoforms Play Distinct Roles in Environmental Adaptation in Rice., 2017, Rice (N Y).
- Genbank accession number
- Key message
- OsPORA was expressed at high levels in developing leaves and decreased dramatically in fully mature leaves, whereas OsPORB expression was relatively constant throughout leaf development, similar to expression patterns of AtPORA and AtPORB in Arabidopsis
- Our results demonstrate that OsPORB is essential for maintaining light-dependent Chl synthesis throughout leaf development, especially under HL conditions, whereas OsPORA mainly functions in the early stages of leaf development
- OsPORA is expressed in the dark during early leaf development; OsPORB is expressed throughout leaf development regardless of light conditions
- The physiological function of OsPORB in response to constant light or during reproductive growth cannot be completely replaced by constitutive activity of OsPORA, although the biochemical functions of OsPORA and OsPORB are redundant
- Connection
- OsNOA1, OsPORA, NOA1 functions in a temperature-dependent manner to regulate chlorophyll biosynthesis and Rubisco formation in rice, Further expression analyses identified several candidate genes, including OsPorA (NADPH: protochlorophyllide oxidoreductase A), OsrbcL (Rubisco large subunit), OsRALyase (Ribosomal RNA apurinic site specific lyase) and OsPuf4 (RNA-binding protein of the Puf family), which may be involved in OsNOA1-regulated chlorophyll biosynthesis and Rubisco formation
- OsPORA, OsPuf4, NOA1 functions in a temperature-dependent manner to regulate chlorophyll biosynthesis and Rubisco formation in rice, Further expression analyses identified several candidate genes, including OsPorA (NADPH: protochlorophyllide oxidoreductase A), OsrbcL (Rubisco large subunit), OsRALyase (Ribosomal RNA apurinic site specific lyase) and OsPuf4 (RNA-binding protein of the Puf family), which may be involved in OsNOA1-regulated chlorophyll biosynthesis and Rubisco formation
- OsPORA, OsRALyase, NOA1 functions in a temperature-dependent manner to regulate chlorophyll biosynthesis and Rubisco formation in rice, Further expression analyses identified several candidate genes, including OsPorA (NADPH: protochlorophyllide oxidoreductase A), OsrbcL (Rubisco large subunit), OsRALyase (Ribosomal RNA apurinic site specific lyase) and OsPuf4 (RNA-binding protein of the Puf family), which may be involved in OsNOA1-regulated chlorophyll biosynthesis and Rubisco formation
- OsPORA, OsPORB~FGL, The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions, OsPORA expression is repressed by light and OsPORB expression is regulated in a circadian rhythm in short-day conditions
- OsPORA, OsPORB~FGL, The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions, OsPORA was expressed at high levels in developing leaves and decreased dramatically in fully mature leaves, whereas OsPORB expression was relatively constant throughout leaf development, similar to expression patterns of AtPORA and AtPORB in Arabidopsis
- OsPORA, OsPORB~FGL, The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions, Our results demonstrate that OsPORB is essential for maintaining light-dependent Chl synthesis throughout leaf development, especially under HL conditions, whereas OsPORA mainly functions in the early stages of leaf development
- OsPORA, YLC1~OsV5A, Differential Regulation of Protochlorophyllide Oxidoreductase Abundances by VIRESCENT 5A OsV5A and VIRESCENT 5B OsV5B in Rice Seedlings., OsV5A and OsV5B interact with two rice PORs (OsPORA and OsPORB) inside chloroplasts and they stabilize OsPORB in vitro under oxidative stress
- OsPORA, OsV5B, Differential Regulation of Protochlorophyllide Oxidoreductase Abundances by VIRESCENT 5A OsV5A and VIRESCENT 5B OsV5B in Rice Seedlings., OsV5A and OsV5B interact with two rice PORs (OsPORA and OsPORB) inside chloroplasts and they stabilize OsPORB in vitro under oxidative stress
- OsPORA, OsPORB~FGL, Two NADPH: Protochlorophyllide Oxidoreductase POR Isoforms Play Distinct Roles in Environmental Adaptation in Rice., Rice has two POR isoforms, OsPORA and OsPORB
- OsPORA, OsPORB~FGL, Two NADPH: Protochlorophyllide Oxidoreductase POR Isoforms Play Distinct Roles in Environmental Adaptation in Rice., OsPORA is expressed in the dark during early leaf development; OsPORB is expressed throughout leaf development regardless of light conditions
- OsPORA, OsPORB~FGL, Two NADPH: Protochlorophyllide Oxidoreductase POR Isoforms Play Distinct Roles in Environmental Adaptation in Rice., To investigate whether the function of OsPORA can complement that of OsPORB, we constitutively overexpressed OsPORA in fgl mutant
- OsPORA, OsPORB~FGL, Two NADPH: Protochlorophyllide Oxidoreductase POR Isoforms Play Distinct Roles in Environmental Adaptation in Rice., The physiological function of OsPORB in response to constant light or during reproductive growth cannot be completely replaced by constitutive activity of OsPORA, although the biochemical functions of OsPORA and OsPORB are redundant
Prev Next