- Information
- Symbol: OsPuf4
- MSU: LOC_Os12g30520
- RAPdb: Os12g0488900
- PSP score
- LOC_Os12g30520.1: 0.8587
- PLAAC score
- LOC_Os12g30520.1: 0
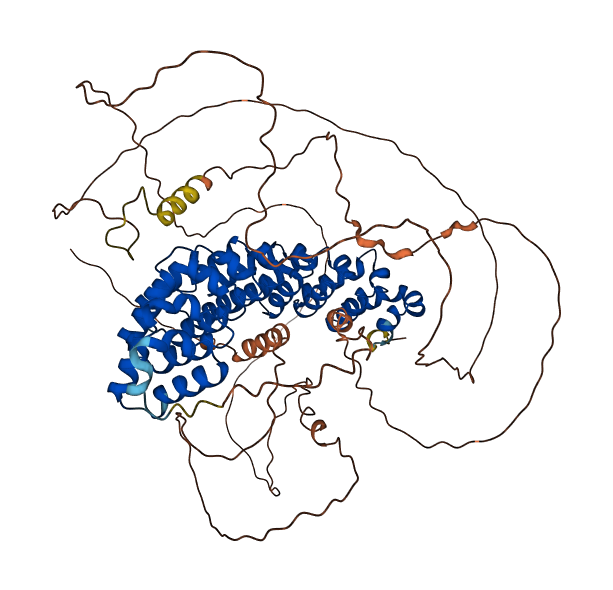
- pLDDT score
- 59.85
- Protein Structure from AlphaFold and UniProt
 )
)
- MolPhase score
- LOC_Os12g30520.1: 0.8587
- MolPhaseResult
- Publication
- Genbank accession number
-
Key message
- Connection
- OsPORA, OsPuf4, NOA1 functions in a temperature-dependent manner to regulate chlorophyll biosynthesis and Rubisco formation in rice, Further expression analyses identified several candidate genes, including OsPorA (NADPH: protochlorophyllide oxidoreductase A), OsrbcL (Rubisco large subunit), OsRALyase (Ribosomal RNA apurinic site specific lyase) and OsPuf4 (RNA-binding protein of the Puf family), which may be involved in OsNOA1-regulated chlorophyll biosynthesis and Rubisco formation
- OsNOA1, OsPuf4, NOA1 functions in a temperature-dependent manner to regulate chlorophyll biosynthesis and Rubisco formation in rice, Further expression analyses identified several candidate genes, including OsPorA (NADPH: protochlorophyllide oxidoreductase A), OsrbcL (Rubisco large subunit), OsRALyase (Ribosomal RNA apurinic site specific lyase) and OsPuf4 (RNA-binding protein of the Puf family), which may be involved in OsNOA1-regulated chlorophyll biosynthesis and Rubisco formation
- OsPuf4, OsRALyase, NOA1 functions in a temperature-dependent manner to regulate chlorophyll biosynthesis and Rubisco formation in rice, Further expression analyses identified several candidate genes, including OsPorA (NADPH: protochlorophyllide oxidoreductase A), OsrbcL (Rubisco large subunit), OsRALyase (Ribosomal RNA apurinic site specific lyase) and OsPuf4 (RNA-binding protein of the Puf family), which may be involved in OsNOA1-regulated chlorophyll biosynthesis and Rubisco formation
Prev Next
