- Information
- Symbol: OsRALyase
- MSU: LOC_Os02g29150
- RAPdb: Os02g0493100
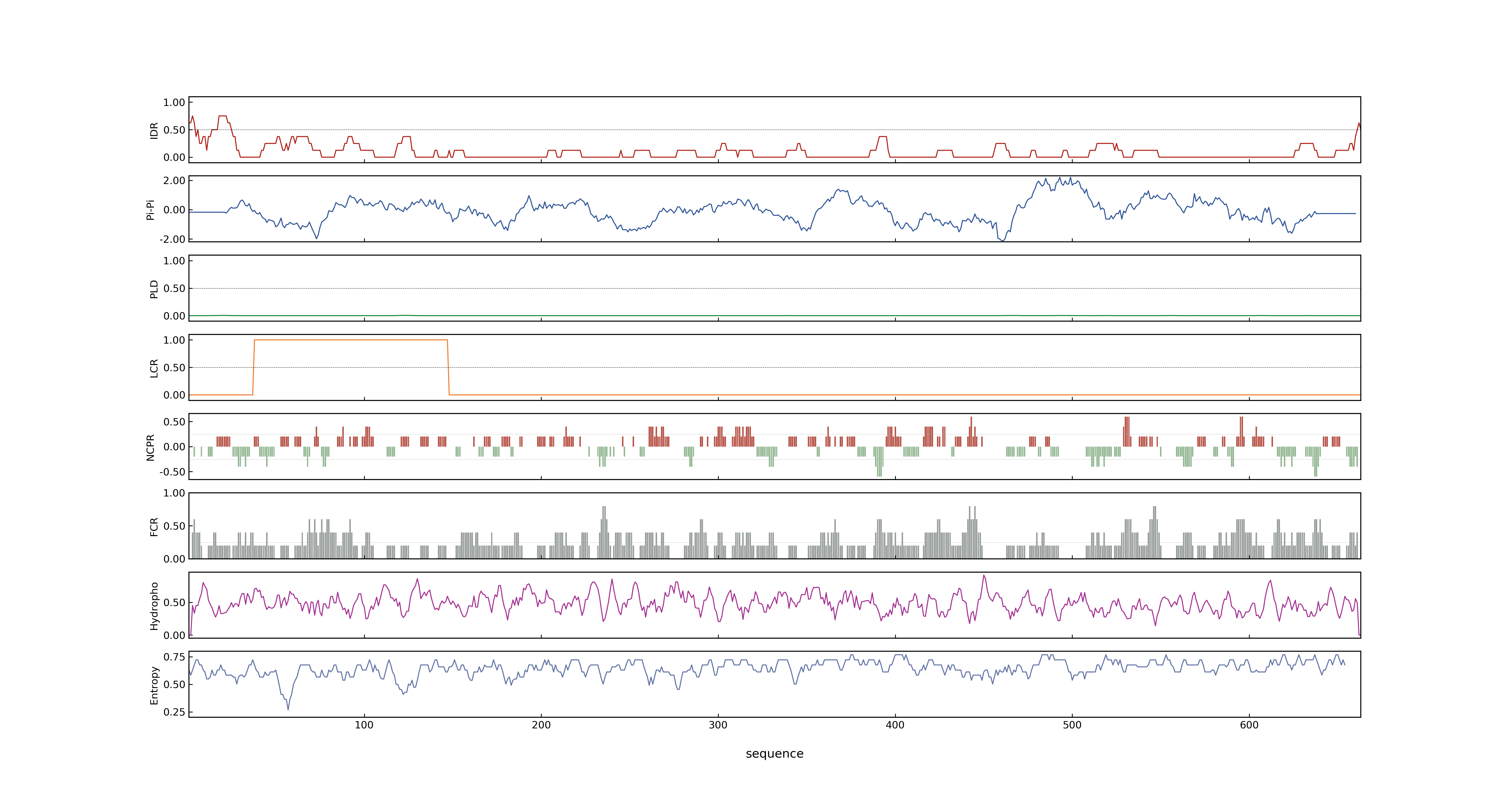
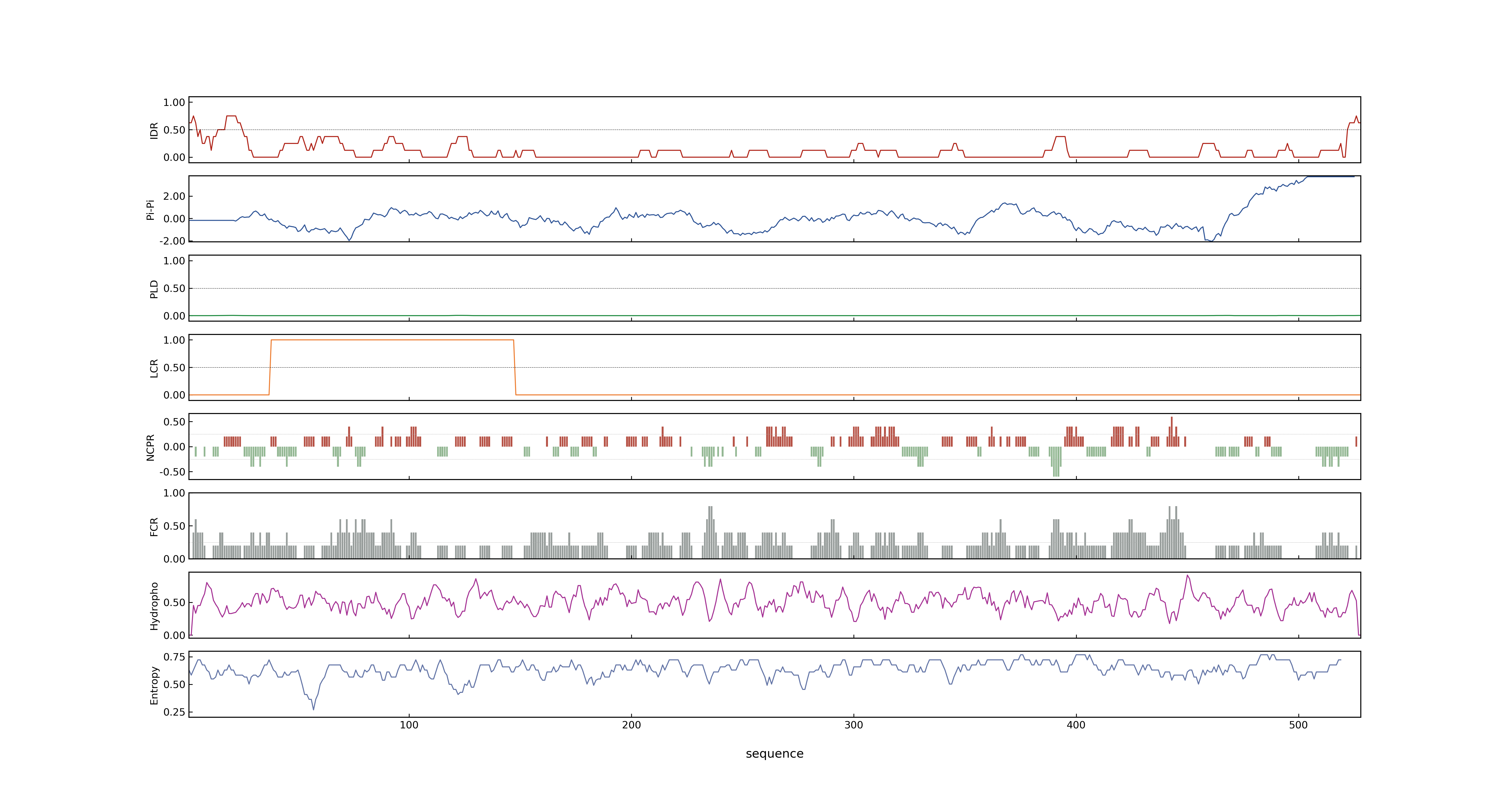
- PSP score
- LOC_Os02g29150.2: 0.0121
- LOC_Os02g29150.1: 0.0649
- PLAAC score
- LOC_Os02g29150.2: 0
- LOC_Os02g29150.1: 0
- pLDDT score
- NA
- MolPhase score
- LOC_Os02g29150.1: 0.05084630
- LOC_Os02g29150.2: 0.35747680
- MolPhase Result
- Publication
-
Genbank accession number
-
Key message
- Connection
- OsNOA1, OsRALyase, NOA1 functions in a temperature-dependent manner to regulate chlorophyll biosynthesis and Rubisco formation in rice, Further expression analyses identified several candidate genes, including OsPorA (NADPH: protochlorophyllide oxidoreductase A), OsrbcL (Rubisco large subunit), OsRALyase (Ribosomal RNA apurinic site specific lyase) and OsPuf4 (RNA-binding protein of the Puf family), which may be involved in OsNOA1-regulated chlorophyll biosynthesis and Rubisco formation
- OsPORA, OsRALyase, NOA1 functions in a temperature-dependent manner to regulate chlorophyll biosynthesis and Rubisco formation in rice, Further expression analyses identified several candidate genes, including OsPorA (NADPH: protochlorophyllide oxidoreductase A), OsrbcL (Rubisco large subunit), OsRALyase (Ribosomal RNA apurinic site specific lyase) and OsPuf4 (RNA-binding protein of the Puf family), which may be involved in OsNOA1-regulated chlorophyll biosynthesis and Rubisco formation
- OsPuf4, OsRALyase, NOA1 functions in a temperature-dependent manner to regulate chlorophyll biosynthesis and Rubisco formation in rice, Further expression analyses identified several candidate genes, including OsPorA (NADPH: protochlorophyllide oxidoreductase A), OsrbcL (Rubisco large subunit), OsRALyase (Ribosomal RNA apurinic site specific lyase) and OsPuf4 (RNA-binding protein of the Puf family), which may be involved in OsNOA1-regulated chlorophyll biosynthesis and Rubisco formation
Prev Next