- Information
- Symbol: OsRMC,OsRLK
- MSU: LOC_Os04g56430
- RAPdb: Os04g0659300
- PSP score
- LOC_Os04g56430.1: 0.0666
- PLAAC score
- LOC_Os04g56430.1: 0
- pLDDT score
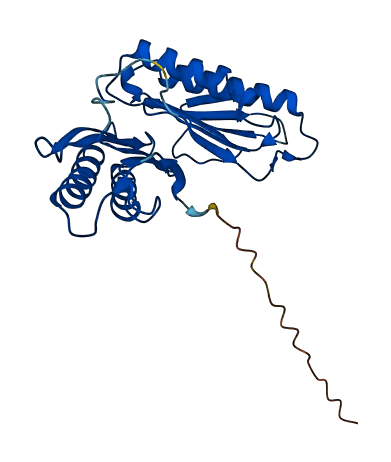
- 91.31
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os04g56430.1: 0.00147224
- MolPhase Result
- Publication
- OsRMC, a negative regulator of salt stress response in rice, is regulated by two AP2/ERF transcription factors, 2013, Plant Mol Biol.
- RNAi knockdown of Oryza sativa root meander curling gene led to altered root development and coiling which were mediated by jasmonic acid signalling in rice, 2007, Plant Cell Environ.
- A receptor-like protein RMC is involved in regulation of iron acquisition in rice, 2013, J Exp Bot.
- Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis, 2009, Plant Physiol.
- Genbank accession number
- Key message
- Our results suggest that OsRMC of the DUF26 subfamily involved in JA signal transduction mediates root development and negatively regulates root curling in rice
- Among these, we have chosen OsRMC to study its transcriptional regulation in rice seedlings subjected to high salinity
- To investigate how OsRMC is regulated in response to high salinity, a salt-induced rice cDNA expression library was constructed and subsequently screened using the yeast one-hybrid system and the OsRMC promoter as bait
- The results show that knocking down the expression level of OsRMC in transgenic rice led to insensitive seed germination, enhanced growth inhibition, and improved salt stress tolerance to NaCl than in untransgenic plants
- OsRMC, a negative regulator of salt stress response in rice, is regulated by two AP2/ERF transcription factors
- Thus, this gene was renamed Oryza sativa root meander curling (OsRMC)
- Here, we evaluated the role of a previously clarified gene encoding a receptor-like protein from rice, OsRMC, in the regulation of Fe acquisition by comparing Fe concentration, biomass, and expression patterns of genes associated with Fe mobilization and transport in wild-type (WT) rice with those in OsRMC overexpression and RNA interference (RNAi) knockdown transgenic rice plants
- sativa root meander curling (OsRMC), has shown drastically increased abundance in response to salt stress during the initial phase
- OsRMC RNA interference transgenic rice has been generated to assess the function of OsRMC in the salt stress response
- Here, we describe a JA-induced putative receptor-like protein (OsRLK, AAL87185) functioning in root development in rice
- Expression of OsRMC was upregulated in both shoots and roots upon exposure of WT to Fe-deficient medium
- Expression levels of OsRMC were positively correlated with Fe concentration in rice plants under both Fe-sufficient and Fe-deficient conditions such that overexpression and RNAi lines had higher and lower Fe concentration in both roots and shoots than WT plants, respectively
- OsRMC may also play a role in regulation of Fe deficiency-induced changes in root growth, as evidenced by greater and smaller root systems of OsRMC overexpression lines and RNAi lines than WT under Fe-deficient conditions, respectively
- These novel findings highlight an important role of OsRMC played in mediation of Fe acquisition and root growth in rice, particularly under Fe-deficient conditions
- Moreover, overexpression of OsRMC conferred greater accumulation of Fe in mature seeds under Fe-sufficient conditions
- OsRMC encodes a receptor-like kinase described as a negative regulator of salt stress responses in rice
- Connection
- OsIRO2, OsRMC~OsRLK, A receptor-like protein RMC is involved in regulation of iron acquisition in rice, Several Fe deficiency-responsive genes including OsDMAS1, OsNAS1, OsNAS2, OsNAAT1, OsIRT1, OsYSL15, and OsIRO2 were up- and downregulated in OsRMC-overexpressing and RNAi plants compared with WT rice plants
- OsIRT1, OsRMC~OsRLK, A receptor-like protein RMC is involved in regulation of iron acquisition in rice, Several Fe deficiency-responsive genes including OsDMAS1, OsNAS1, OsNAS2, OsNAAT1, OsIRT1, OsYSL15, and OsIRO2 were up- and downregulated in OsRMC-overexpressing and RNAi plants compared with WT rice plants
- OsNAAT1, OsRMC~OsRLK, A receptor-like protein RMC is involved in regulation of iron acquisition in rice, Several Fe deficiency-responsive genes including OsDMAS1, OsNAS1, OsNAS2, OsNAAT1, OsIRT1, OsYSL15, and OsIRO2 were up- and downregulated in OsRMC-overexpressing and RNAi plants compared with WT rice plants
- OsNAS1, OsRMC~OsRLK, A receptor-like protein RMC is involved in regulation of iron acquisition in rice, Several Fe deficiency-responsive genes including OsDMAS1, OsNAS1, OsNAS2, OsNAAT1, OsIRT1, OsYSL15, and OsIRO2 were up- and downregulated in OsRMC-overexpressing and RNAi plants compared with WT rice plants
- OsNAS2, OsRMC~OsRLK, A receptor-like protein RMC is involved in regulation of iron acquisition in rice, Several Fe deficiency-responsive genes including OsDMAS1, OsNAS1, OsNAS2, OsNAAT1, OsIRT1, OsYSL15, and OsIRO2 were up- and downregulated in OsRMC-overexpressing and RNAi plants compared with WT rice plants
- OsRMC~OsRLK, OsYSL15, A receptor-like protein RMC is involved in regulation of iron acquisition in rice, Several Fe deficiency-responsive genes including OsDMAS1, OsNAS1, OsNAS2, OsNAAT1, OsIRT1, OsYSL15, and OsIRO2 were up- and downregulated in OsRMC-overexpressing and RNAi plants compared with WT rice plants
- OsHOS1, OsRMC~OsRLK, The rice E3 ubiquitin ligase OsHOS1 modulates the expression of OsRMC, a gene involved in root mechano-sensing, through the interaction with two ERF transcription factors., The rice E3 ubiquitin ligase OsHOS1 modulates the expression of OsRMC, a gene involved in root mechano-sensing, through the interaction with two ERF transcription factors.
- OsHOS1, OsRMC~OsRLK, The rice E3 ubiquitin ligase OsHOS1 modulates the expression of OsRMC, a gene involved in root mechano-sensing, through the interaction with two ERF transcription factors., Using the yeast two-hybrid system and BiFC assays we showed that OsHOS1 interacts with two ERF transcription factors (TFs), OsEREBP1 and OsEREBP2, known to regulate OsRMC gene expression
Prev Next