- Information
- Symbol: OsRNS4,OsRRP
- MSU: LOC_Os09g36680
- RAPdb: Os09g0537700
- PSP score
- LOC_Os09g36680.1: 0.4892
- PLAAC score
- LOC_Os09g36680.1: 0
- pLDDT score
- 88.54
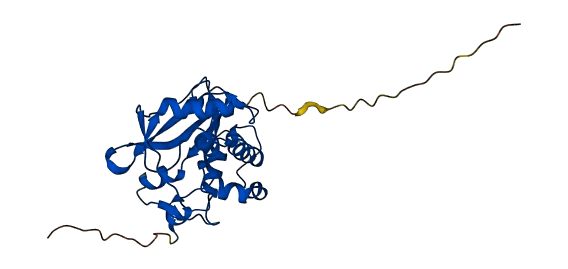
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os09g36680.1: 0.00424213
- MolPhase Result
- Publication
- Overexpression of an S-like ribonuclease gene, OsRNS4, confers enhanced tolerance to high salinity and hyposensitivity to phytochrome-mediated light signals in rice, 2014, Plant Sci.
- Cloning and characterization of an RNase-related protein gene preferentially expressed in rice stems, 2006, Biosci Biotechnol Biochem.
- Genbank accession number
- Key message
- OsRRP is preferentially expressed in stems of wild-type rice and is significantly down-regulated in an increased tillering dwarf mutant ext37
- The seedlings overexpressing OsRNS4 had longer coleoptiles and first leaves than wild-type seedlings under red light (R) and far-red light (FR), suggesting negative regulation of OsRNS4 in photomorphogenesis in rice seedlings
- Moreover, ABA-induced growth inhibition of rice seedlings was significantly increased in the OsRNS4-overexpression (OsRNS4-OX) lines compared with that in WT, suggesting that OsRNS4 probably acts as a positive regulator in ABA responses in rice seedlings
- Here, we investigated the expression patterns and roles of an S-like RNase gene, OsRNS4, in abscisic acid (ABA)-mediated responses and phytochrome-mediated light responses as well as salinity tolerance in rice
- In addition, our results demonstrate that OsRNS4-OX lines have enhanced tolerance to high salinity compared to WT
- Overexpression of an S-like ribonuclease gene, OsRNS4, confers enhanced tolerance to high salinity and hyposensitivity to phytochrome-mediated light signals in rice
- OsRNS4 expression was regulated by salt, PEG and ABA
- Connection
Prev Next