- Information
- Symbol: OsYUCCA1,OsYUC1
- MSU: LOC_Os01g45760
- RAPdb: Os01g0645400
- PSP score
- LOC_Os01g45760.2: 0.0091
- LOC_Os01g45760.1: 0.006
- PLAAC score
- LOC_Os01g45760.2: 0
- LOC_Os01g45760.1: 0
- pLDDT score
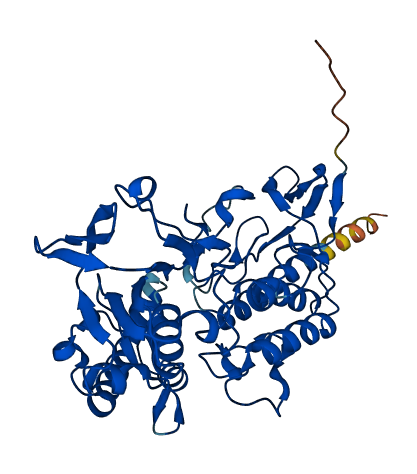
- 93.04
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os01g45760.1: 0.00950465
- LOC_Os01g45760.2: 0.00393054
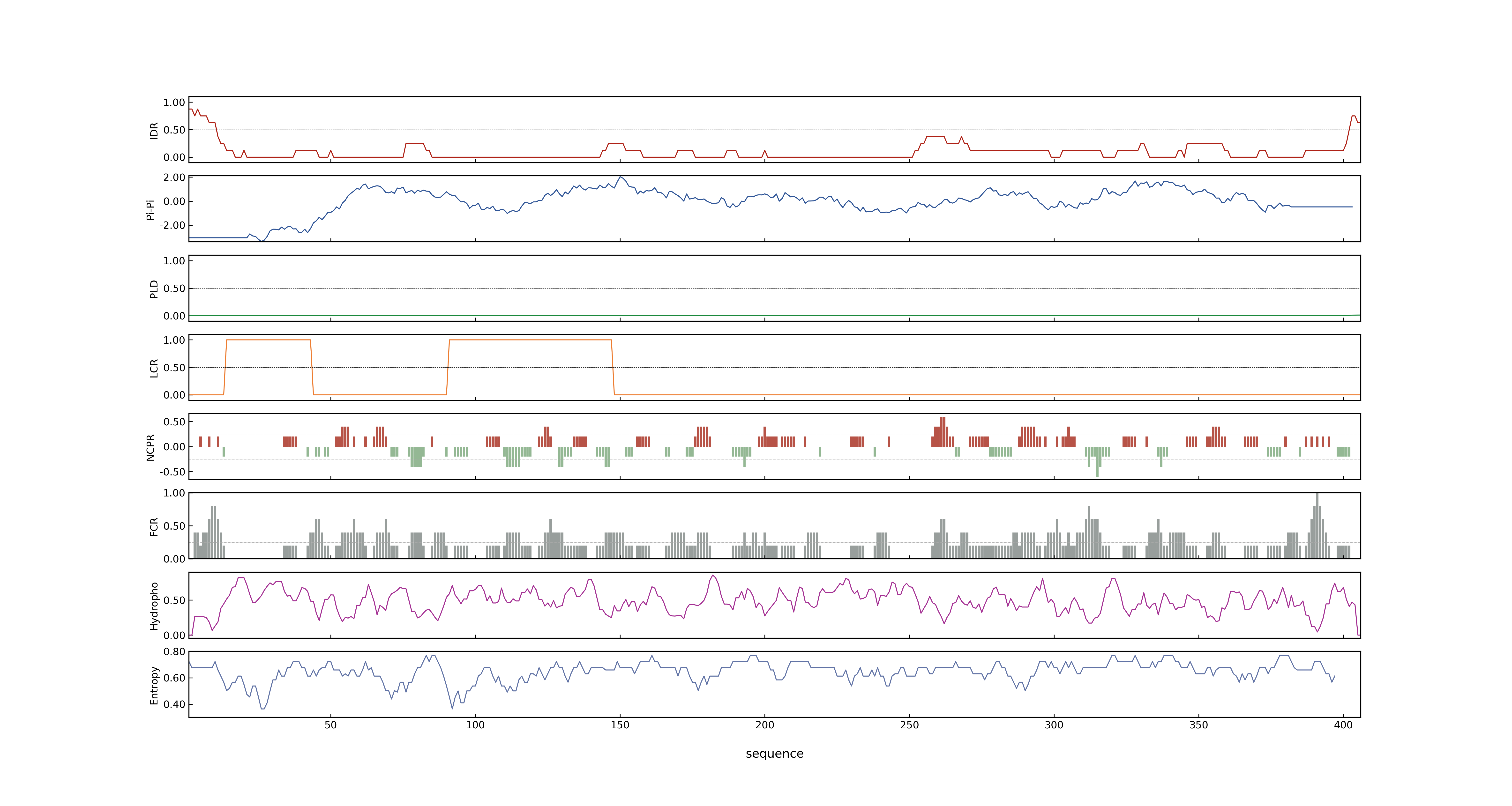
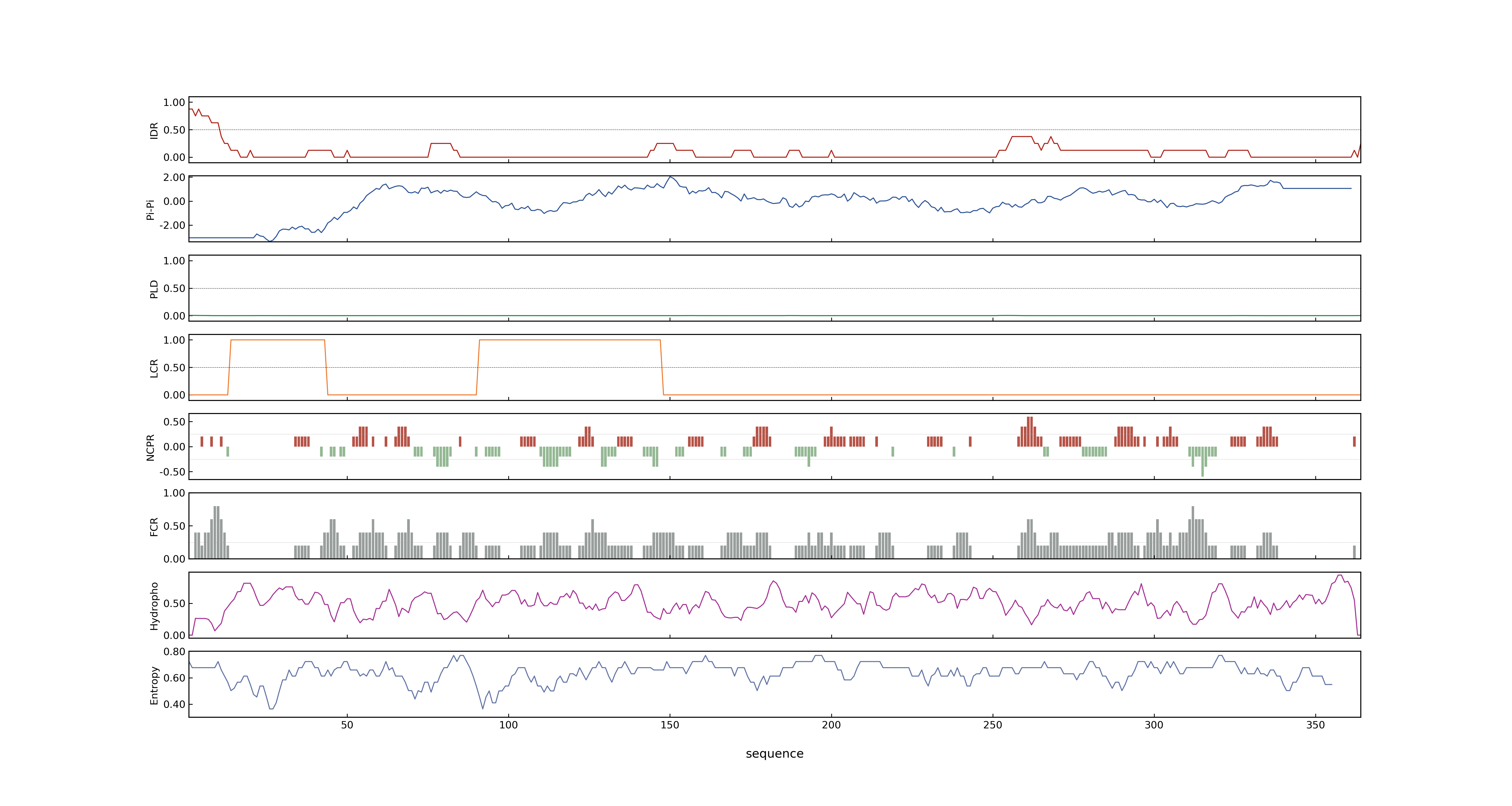
- MolPhase Result
- Publication
- A role for a dioxygenase in auxin metabolism and reproductive development in rice, 2013, Dev Cell.
- Cloning, characterization and expression of OsFMOt in rice encoding a flavin monooxygenase, 2013, J Genet.
- A rice tryptophan deficient dwarf mutant, tdd1, contains a reduced level of indole acetic acid and develops abnormal flowers and organless embryos, 2009, Plant J.
- Auxin biosynthesis by the YUCCA genes in rice, 2007, Plant Physiol.
- Genbank accession number
- Key message
- Furthermore, exogenous application of IAA or overexpression of the auxin biosynthesis gene OsYUCCA1 phenocopies the dao mutants
- To investigate the molecular mechanisms of IAA synthesis in rice (Oryza sativa), we identified seven YUCCA-like genes (named OsYUCCA1-7) in the rice genome
- Plants overexpressing OsYUCCA1 exhibited increased IAA levels and characteristic auxin overproduction phenotypes, whereas plants expressing antisense OsYUCCA1 cDNA displayed defects that are similar to those of rice auxin-insensitive mutants
- These observations are consistent with an important role for the rice enzyme OsYUCCA1 in IAA biosynthesis via the tryptophan-dependent pathway
- OsYUCCA1 was expressed in almost all of the organs tested, but its expression was restricted to discrete areas, including the tips of leaves, roots, and vascular tissues, where it overlapped with expression of a beta-glucuronidase reporter gene controlled by the auxin-responsive DR5 promoter
- Trp feeding completely rescued the mutant phenotypes and moderate expression of OsYUCCA1, which encodes a key enzyme in Trp-dependent IAA biosynthesis, also rescued plant height and root length, indicating that the abnormal phenotypes of tdd1 are caused predominantly by Trp and IAA deficiency
- Connection
- DAO, OsYUCCA1~OsYUC1, A role for a dioxygenase in auxin metabolism and reproductive development in rice, Furthermore, exogenous application of IAA or overexpression of the auxin biosynthesis gene OsYUCCA1 phenocopies the dao mutants
- OsFMO~OsCOW1~NAL7, OsYUCCA1~OsYUC1, Cloning, characterization and expression of OsFMOt in rice encoding a flavin monooxygenase, OsFMO(t) showed high amino acid sequence identity with FMO proteins from other plants, in particular with YUCCA from Arabidopsis, FLOOZY from Petunia, and OsYUCCA1 from rice
- OsYUCCA1~OsYUC1, TDD1~OASB1, A rice tryptophan deficient dwarf mutant, tdd1, contains a reduced level of indole acetic acid and develops abnormal flowers and organless embryos, Trp feeding completely rescued the mutant phenotypes and moderate expression of OsYUCCA1, which encodes a key enzyme in Trp-dependent IAA biosynthesis, also rescued plant height and root length, indicating that the abnormal phenotypes of tdd1 are caused predominantly by Trp and IAA deficiency
- OsARID3, OsYUCCA1~OsYUC1, OsARID3, an AT-rich Interaction Domain-containing protein, is required for shoot meristem development in rice, Furthermore, our chromatin immunoprecipitation results demonstrate that OsARID3 binds directly to the KNOXI gene OSH71, the auxin biosynthetic genes OsYUC1 and OsYUC6, and the cytokinin biosynthetic genes OsIPT2 and OsIPT7.
- OsARID3, OsYUCCA1~OsYUC1, OsARID3, an AT-rich Interaction Domain-containing protein, is required for shoot meristem development in rice., Furthermore, chromatin immunoprecipitation results demonstrate that OsARID3 binds directly to the KNOXI gene OSH71, the auxin biosynthetic genes OsYUC1 and OsYUC6, and the cytokinin biosynthetic genes OsIPT2 and OsIPT7
Prev Next