- Information
- Symbol: Osmyb4
- MSU: LOC_Os04g43680
- RAPdb: Os04g0517100
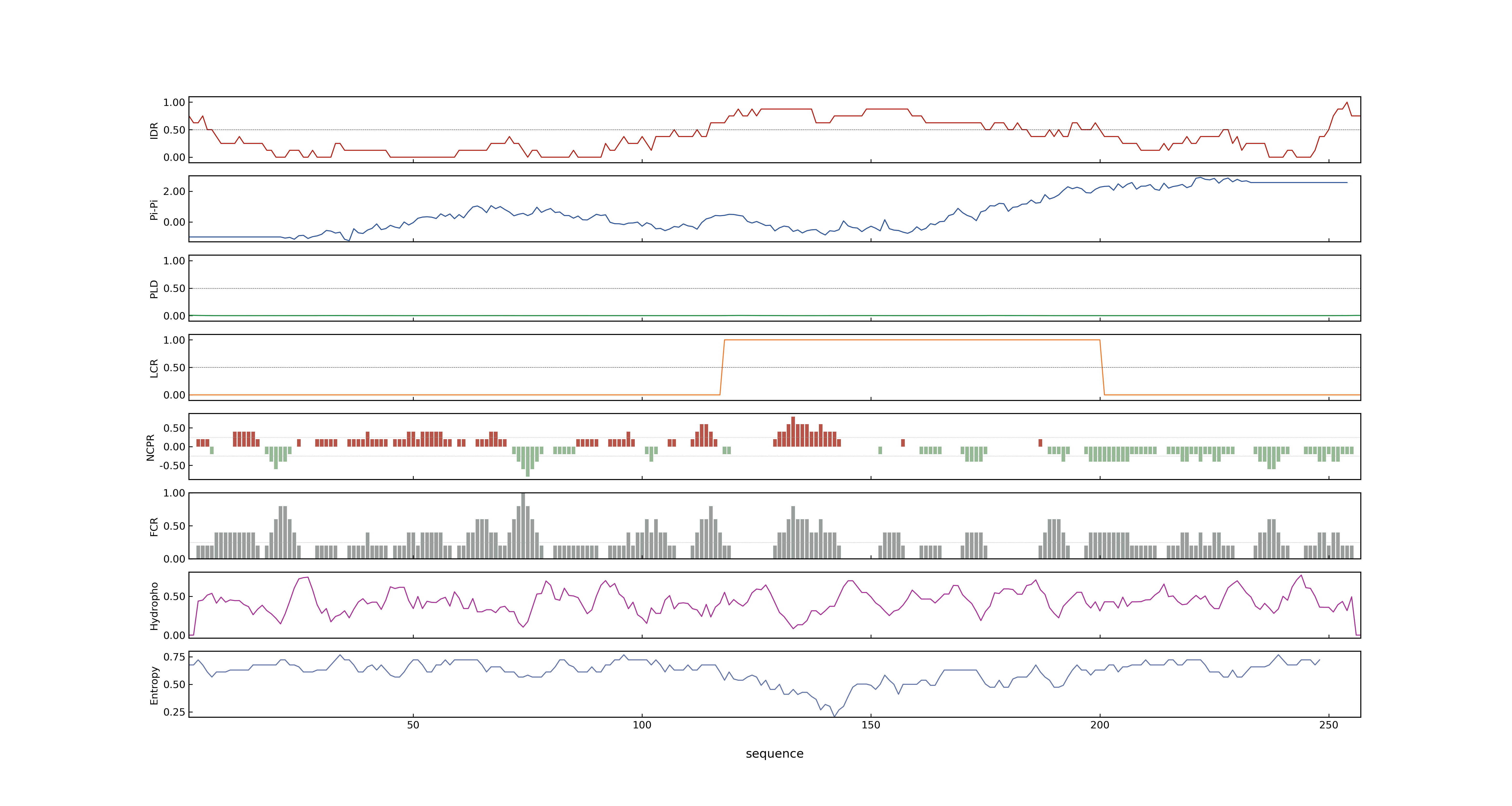
- PSP score
- LOC_Os04g43680.1: 0.2285
- PLAAC score
- LOC_Os04g43680.1: 0
- pLDDT score
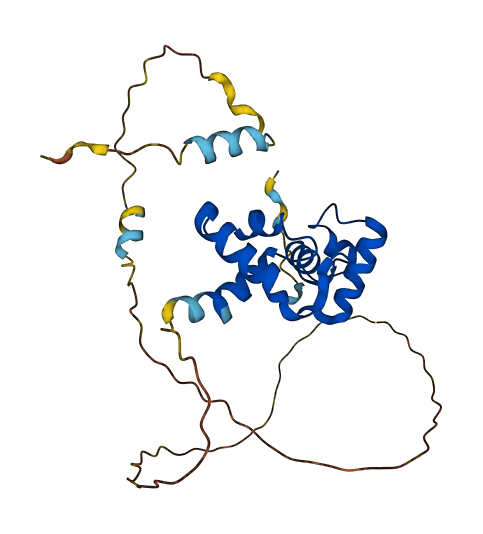
- 69.94
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os04g43680.1: 0.98955088
- MolPhase Result
- Publication
- Evaluation of transgenic tomato plants ectopically expressing the rice Osmyb4 gene, 2007, Plant Science.
- Cloning and expression of five myb-related genes from rice seed, 1997, Gene.
- The ectopic expression of the rice Osmyb4 gene in Arabidopsis increases tolerance to abiotic, environmental and biotic stresses, 2006, Physiological and Molecular Plant Pathology.
- Metabolic response to cold and freezing of Osteospermum ecklonis overexpressing Osmyb4, 2010, Plant Physiol Biochem.
- The rice Osmyb4 gene enhances tolerance to frost and improves germination under unfavourable conditions in transgenic barley plants, 2012, J Appl Genet.
- Response of transgenic rape plants bearing the Osmyb4 gene from rice encoding a trans-factor to low above-zero temperature, 2011, Russian Journal of Plant Physiology.
- Supra-optimal expression of the cold-regulated OsMyb4 transcription factor in transgenic rice changes the complexity of transcriptional network with major effects on stress tolerance and panicle development, 2010, Plant Cell Environ.
- Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants, 2004, Plant J.
- OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice, 2014, Plant Mol Biol.
- Homotypic clustering of OsMYB4 binding site motifs in promoters of the rice genome and cellular-level implications on sheath blight disease resistance., 2015, Gene.
- Genbank accession number
- Key message
- Like Arabidopsis, tomato plants overexpressing Osmyb4 acquired a higher tolerance to drought stress and to virus disease
- Real-time PCR analysis revealed that the abiotic stress-related genes, OsVHA-B, OsNHX1, COR413-TM1, and OsMyb4, were up-regulated in overexpressing lines, while these same genes were down-regulated in RNAi lines
- The Osmyb4 rice gene, coding for a transcription factor, proved to be efficient against different abiotic stresses as a trans(cis)gene in several plant species, although the effectiveness was dependent on the host genomic background
- These results provide evidence that the cold dependent expression of Osmyb4 can efficiently improved frost tolerance and germination vigour at low temperature without deleterious effect on plant growth
- Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants
- In seed, the mRNA levels of Osmyb1 and Osmyb4 genes reached a maximum at 14 days after flowering (DAF), suggesting that these genes may play a role in seed maturation
- The R2R3-type OsMyb4 transcription factor of rice has been shown to play a role in the regulation of osmotic adjustment in heterologous overexpression studies
- Supra-optimal expression of the cold-regulated OsMyb4 transcription factor in transgenic rice changes the complexity of transcriptional network with major effects on stress tolerance and panicle development
- OsMyb4 network was dissected based on commonalities between the global chilling stress transcriptome and the transcriptome configured by OsMyb4 overexpression
- Response of transgenic rape plants bearing the Osmyb4 gene from rice encoding a trans-factor to low above-zero temperature
- The Osmyb4 rice gene encodes a Myb transcription factor involved in cold acclimation
- Finally, in Osmyb4 transgenic plants, the expression of genes participating in different cold-induced pathways is affected, suggesting that Myb4 represents a master switch in cold tolerance
- OsMyb4 network is independent of drought response element binding protein/C-repeat binding factor (DREB/CBF) and its sub-regulons operate with possible co-regulators including nuclear factor-Y
- The ectopic expression of the rice Osmyb4 gene in Arabidopsis increases tolerance to abiotic, environmental and biotic stresses
- The rice Osmyb4 gene, coding for a MYB transcription factor, is expressed at low levels in rice coleoptiles under normal conditions and strongly induced at 4 ÃÂÃÂÃÂÃÂÃÂÃÂÃÂÃÂÃÂÃÂÃÂÃÂÃÂÃÂÃÂðC
- In this study, we explored the potential of Osmyb4 to enhance the cold and freezing tolerance of Osteospermum ecklonis, an ornamental and perennial plant native to South Africa, because of an increasing interest in growing this species in Europe where winter temperatures are low
- OsMyb4 controls a hierarchical network comprised of several regulatory sub-clusters associated with cellular defense and rescue, metabolism and development
- The constitutive expression of the rice Osmyb4 gene in Arabidopsis plants gives rise to enhanced abiotic and biotic stress tolerance, probably by activating several stress-inducible pathways
- Over-expression of Osmyb4 cDNA in rice leaf tissues by agro-infection failed to result in similar over-expression of aminotransferase, ankyrin and WRKY 12 as expected
- Homotypic clustering of OsMYB4 binding site motifs in promoters of the rice genome and cellular-level implications on sheath blight disease resistance.
- Although the role of OsMYB4 in sheath blight resistance was found to be definitive based on our initial results, artificial over-expression of this TF was observed to be insufficient in regulating the disease resistance related genes
- Among the downstream genes of these promoters, five genes which are known to have a role in disease resistance were selected and the binding capacity of OsMYB4 protein in the promoter regions was analyzed by docking studies
- Connection
- Osmyb1, Osmyb4, Cloning and expression of five myb-related genes from rice seed, In seed, the mRNA levels of Osmyb1 and Osmyb4 genes reached a maximum at 14 days after flowering (DAF), suggesting that these genes may play a role in seed maturation
- Osmyb4, OsNHX1, OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice, Real-time PCR analysis revealed that the abiotic stress-related genes, OsVHA-B, OsNHX1, COR413-TM1, and OsMyb4, were up-regulated in overexpressing lines, while these same genes were down-regulated in RNAi lines
Prev Next