- Information
- Symbol: PHYB,OsphyB
- MSU: LOC_Os03g19590
- RAPdb: Os03g0309200
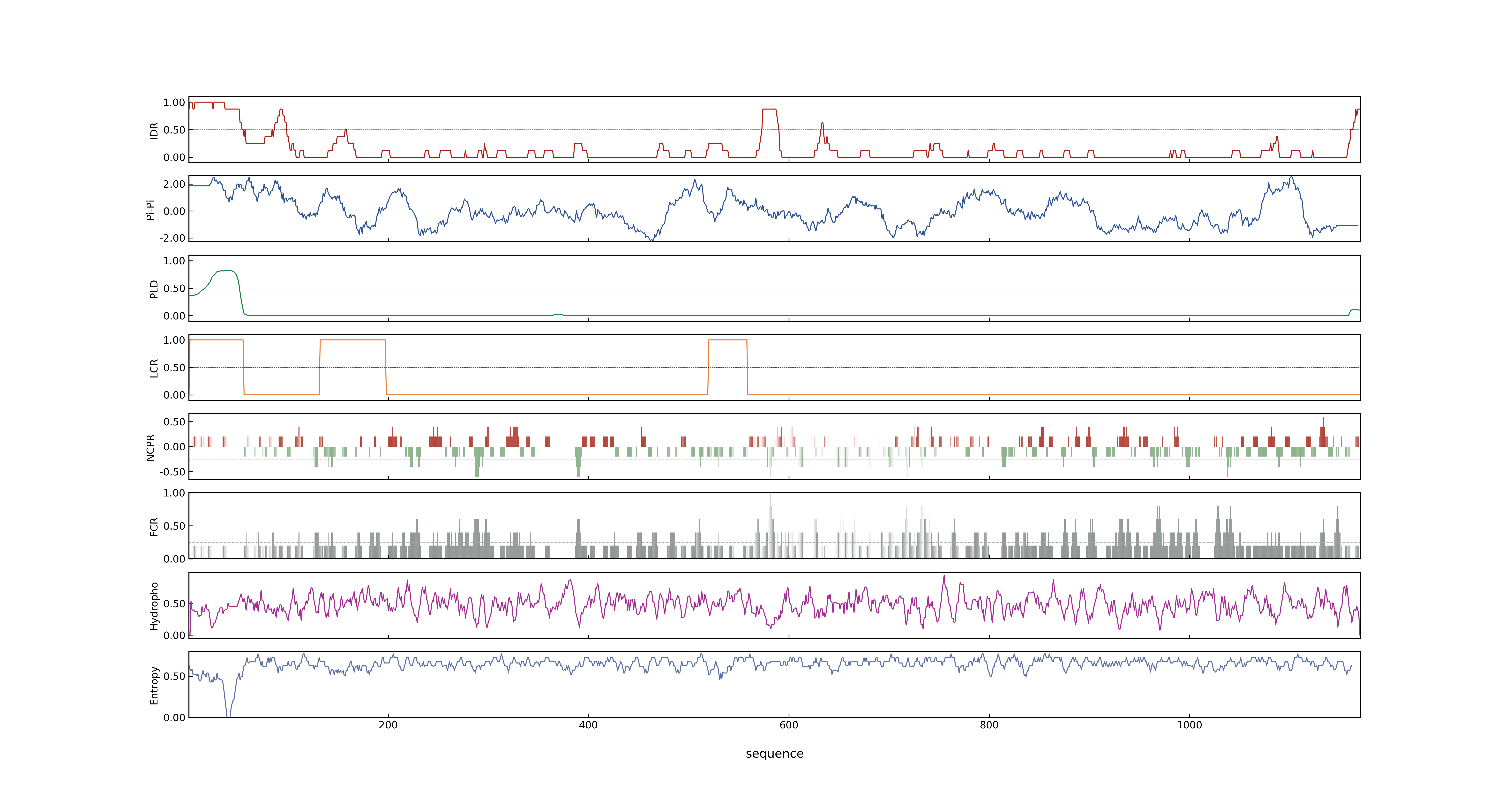
- PSP score
- LOC_Os03g19590.1: 0.3805
- PLAAC score
- LOC_Os03g19590.1: 2.006
(Zheng et al., New Phytol, 2019, 224:306-320)
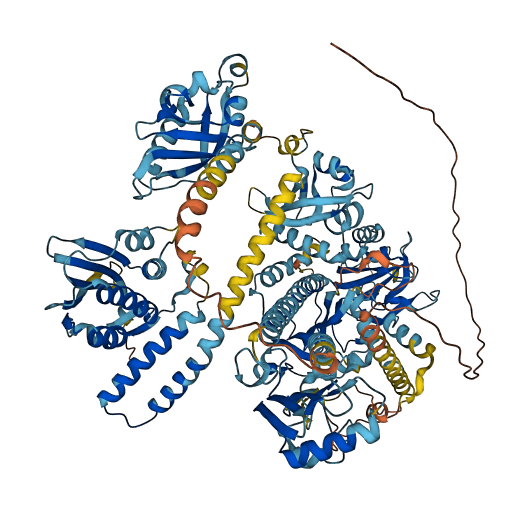
- pLDDT score
- 76.82
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os03g19590.1: 0.99713301
- MolPhase Result
- Publication
- Phytochrome B regulates Heading date 1 Hd1-mediated expression of rice florigen Hd3a and critical day length in rice, 2011, Mol Genet Genomics.
- Phytochrome B control of total leaf area and stomatal density affects drought tolerance in rice, 2012, Plant Mol Biol.
- Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice, 2011, Plant Physiol.
- The multiple contributions of phytochromes to the control of internode elongation in rice, 2011, Plant Physiol.
- A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice, 2009, Development.
- Cryptochrome and phytochrome cooperatively but independently reduce active gibberellin content in rice seedlings under light irradiation, 2012, Plant Cell Physiol.
- Rice JASMONATE RESISTANT 1 is involved in phytochrome and jasmonate signalling, 2008, Plant Cell Environ.
- OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB, 2010, Plant J.
- Footprints of natural and artificial selection for photoperiod pathway genes in Oryza, 2012, Plant J.
- Phytochromes are the sole photoreceptors for perceiving red/far-red light in rice, 2009, Proc Natl Acad Sci U S A.
- Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice, 2005, Plant Cell.
- Comprehensive metabolite profiling of phyA phyB phyC triple mutants to reveal their associated metabolic phenotype in rice leaves, 2009, J Biosci Bioeng.
- Phytochrome B Mediates the Regulation of Chlorophyll Biosynthesis through Transcriptional Regulation of ChlH and GUN4 in Rice Seedlings., 2015, PLoS One.
- Rice Phytochrome B OsPhyB Negatively Regulates Dark- and Starvation-Induced Leaf Senescence., 2015, Plants (Basel).
- OsPhyB-Mediating Novel Regulatory Pathway for Drought Tolerance in Rice Root Identified by a Global RNA-Seq Transcriptome Analysis of Rice Genes in Response to Water Deficiencies., 2017, Front Plant Sci.
- Genbank accession number
- Key message
- To understand the underlying mechanism by which phyB regulates drought tolerance, we analyzed root growth and water loss from the leaves of phyB mutants
- The root system showed no significant difference between the phyB mutants and WT, suggesting that improved drought tolerance has little relation to root growth
- Rice is a short-day plant, and we found that mutation in either phyB or phyC caused moderate early flowering under the long-day photoperiod, while monogenic phyA mutation had little effect on the flowering time
- The phyA mutation, however, in combination with phyB or phyC mutation caused dramatic early flowering
- A gene for 1-aminocyclopropane-1-carboxylate oxidase (ACO1), which is an ethylene biosynthesis gene contributing to internode elongation, was up-regulated in phyAphyBphyC seedlings
- In addition, the transcription levels of several ethylene- or gibberellin (GA)-related genes were changed in phyAphyBphyC mutants, and measurement of the plant hormone levels indicated low ethylene production and bioactive GA levels in the phyAphyBphyC mutants
- We demonstrate that ethylene induced internode elongation and ACO1 expression in phyAphyBphyC seedlings but not in the wild type and that the presence of bioactive GAs was necessary for these effects
- Moreover, phyB and phyA can affect Ghd7 activity and Early heading date1 (a floral inducer) activity in the network, respectively
- In osphyB mutants, OsCOL4 expression was decreased and osphyB oscol4 double mutants flowered at the same time as the osphyB single mutants, indicating OsCOL4 functions downstream of OsphyB
- We also present evidence for two independent pathways through which OsPhyB controls flowering time
- OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB
- Although phyAphyBphyC phytochrome-null mutants in rice (Oryza sativa) have morphological changes and exhibit internode elongation, even as seedlings, it is unknown how phytochromes contribute to the control of internode elongation
- ACO1 expression was controlled mainly by phyA and phyB, and a histochemical analysis showed that ACO1 expression was localized to the basal parts of leaf sheaths of phyAphyBphyC seedlings, similar to mature wild-type plants at the heading stage, when internode elongation was greatly promoted
- The metabolite profiles indicated high accumulation of amino acids, organic acids, sugars, sugar phosphates, and nucleotides in the leaf blades of phyA phyB phyC triple mutants, especially in the young leaves, compared with those in the WT
- We report that phytochrome B (phyB) mutants exhibit improved drought tolerance compared to wild type (WT) rice (Oryza sativa L
- Considering all these findings, we propose that phyB deficiency causes both reduced total leaf area and reduced transpiration per unit leaf area, which explains the reduced water loss and improved drought tolerance of phyB mutants
- We propose that both positive (OsMADS50 and Ehd1) and negative (Hd1, phyB and Ghd7) regulators of RFT1 form a gene network that regulates LD flowering in rice
- Seedlings of phyB and phyB phyC mutants exhibited a partial loss of sensitivity to continuous red light (Rc) but still showed significant deetiolation responses
- Here, we report that phytochrome B (phyB)-mediated suppression of Hd3a is a primary cause of long-day suppression of flowering in rice, based on the three complementary discoveries
- First, overexpression of Hd1 causes a delay in flowering under SD conditions and this effect requires phyB, suggesting that light modulates Hd1 control of Hd3a transcription
- phyB deficiency promoted the expression of both putative ERECTA family genes and EXPANSIN family genes involved in cell expansion in leaves, thus causing greater epidermal cell expansion in the phyB mutants
- In addition, the developed leaves of phyB mutants displayed larger epidermal cells than WT leaves, resulting in reduced stomatal density
- Reduced stomatal density resulted in reduced transpiration per unit leaf area in the phyB mutants
- However, phyB mutants exhibited reduced total leaf area per plant, which was probably due to a reduction in the total number of cells per leaf caused by enhanced expression of Orysa;KRP1 and Orysa;KRP4 (encoding inhibitors of cyclin-dependent kinase complex activity) in the phyB mutants
- We examined the footprints of natural and artificial selections for four major genes of the photoperiod pathway, namely PHYTOCHROME B (PhyB), HEADING DATE 1 (Hd1), HEADING DATE 3a (Hd3a), and EARLY HEADING DATE 1 (Ehd1), by investigation of the patterns of nucleotide polymorphisms in cultivated and wild rice
- Distinct metabolic profiles between phyA phyB phyC triple mutants and the wild type (WT), as well as those between young and mature leaf blades, could be clearly observed by principal component analysis (PCA)
- These results suggest that phyB mediates the regulation of chlorophyll synthesis through transcriptional regulation of these two genes, whose products exert their action at the branching point of the chlorophyll biosynthesis pathway
- The RT-qPCR analysis revealed that several senescence-associated genes, including OsORE1 and OsEIN3, were significantly up-regulated in osphyB-2 mutants, indicating that OsPhyB also inhibits leaf senescence, like Arabidopsis PhyB
- Unlike previous result, we found that OsPhyB represses the activity of ascorbate peroxidase and catalase mediating reactive oxygen species (ROS) processing machinery required for drought tolerance of roots in soil condition, suggesting the potential significance of remaining uncharacterized candidate genes for manipulating drought tolerance in rice
- Connection
- Hd3a, PHYB~OsphyB, Phytochrome B regulates Heading date 1 Hd1-mediated expression of rice florigen Hd3a and critical day length in rice, Here, we report that phytochrome B (phyB)-mediated suppression of Hd3a is a primary cause of long-day suppression of flowering in rice, based on the three complementary discoveries
- Hd3a, PHYB~OsphyB, Phytochrome B regulates Heading date 1 Hd1-mediated expression of rice florigen Hd3a and critical day length in rice, First, overexpression of Hd1 causes a delay in flowering under SD conditions and this effect requires phyB, suggesting that light modulates Hd1 control of Hd3a transcription
- Hd3a, PHYB~OsphyB, Phytochrome B regulates Heading date 1 Hd1-mediated expression of rice florigen Hd3a and critical day length in rice, These results also suggest that phyB-mediated suppression of Hd3a expression is a component of the molecular mechanism for critical day length in rice
- Hd1, PHYB~OsphyB, Phytochrome B regulates Heading date 1 Hd1-mediated expression of rice florigen Hd3a and critical day length in rice, First, overexpression of Hd1 causes a delay in flowering under SD conditions and this effect requires phyB, suggesting that light modulates Hd1 control of Hd3a transcription
- KRP1, PHYB~OsphyB, Phytochrome B control of total leaf area and stomatal density affects drought tolerance in rice, However, phyB mutants exhibited reduced total leaf area per plant, which was probably due to a reduction in the total number of cells per leaf caused by enhanced expression of Orysa;KRP1 and Orysa;KRP4 (encoding inhibitors of cyclin-dependent kinase complex activity) in the phyB mutants
- Ghd7, PHYB~OsphyB, Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice, Our results show that either phyA alone or a genetic combination of phyB and phyC can induce Ghd7 mRNA, whereas phyB alone causes some reduction in levels of Ghd7 mRNA
- Ghd7, PHYB~OsphyB, Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice, Moreover, phyB and phyA can affect Ghd7 activity and Early heading date1 (a floral inducer) activity in the network, respectively
- PHYA~OsPhyA, PHYB~OsphyB, Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice, Our results show that either phyA alone or a genetic combination of phyB and phyC can induce Ghd7 mRNA, whereas phyB alone causes some reduction in levels of Ghd7 mRNA
- PHYA~OsPhyA, PHYB~OsphyB, Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice, Moreover, phyB and phyA can affect Ghd7 activity and Early heading date1 (a floral inducer) activity in the network, respectively
- PHYB~OsphyB, PHYC, Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice, Our results show that either phyA alone or a genetic combination of phyB and phyC can induce Ghd7 mRNA, whereas phyB alone causes some reduction in levels of Ghd7 mRNA
- OsACO1, PHYB~OsphyB, The multiple contributions of phytochromes to the control of internode elongation in rice, A gene for 1-aminocyclopropane-1-carboxylate oxidase (ACO1), which is an ethylene biosynthesis gene contributing to internode elongation, was up-regulated in phyAphyBphyC seedlings
- OsACO1, PHYB~OsphyB, The multiple contributions of phytochromes to the control of internode elongation in rice, ACO1 expression was controlled mainly by phyA and phyB, and a histochemical analysis showed that ACO1 expression was localized to the basal parts of leaf sheaths of phyAphyBphyC seedlings, similar to mature wild-type plants at the heading stage, when internode elongation was greatly promoted
- OsACO1, PHYB~OsphyB, The multiple contributions of phytochromes to the control of internode elongation in rice, We demonstrate that ethylene induced internode elongation and ACO1 expression in phyAphyBphyC seedlings but not in the wild type and that the presence of bioactive GAs was necessary for these effects
- PHYB~OsphyB, PHYC, The multiple contributions of phytochromes to the control of internode elongation in rice, Although phyAphyBphyC phytochrome-null mutants in rice (Oryza sativa) have morphological changes and exhibit internode elongation, even as seedlings, it is unknown how phytochromes contribute to the control of internode elongation
- PHYB~OsphyB, PHYC, The multiple contributions of phytochromes to the control of internode elongation in rice, A gene for 1-aminocyclopropane-1-carboxylate oxidase (ACO1), which is an ethylene biosynthesis gene contributing to internode elongation, was up-regulated in phyAphyBphyC seedlings
- PHYB~OsphyB, PHYC, The multiple contributions of phytochromes to the control of internode elongation in rice, ACO1 expression was controlled mainly by phyA and phyB, and a histochemical analysis showed that ACO1 expression was localized to the basal parts of leaf sheaths of phyAphyBphyC seedlings, similar to mature wild-type plants at the heading stage, when internode elongation was greatly promoted
- PHYB~OsphyB, PHYC, The multiple contributions of phytochromes to the control of internode elongation in rice, In addition, the transcription levels of several ethylene- or gibberellin (GA)-related genes were changed in phyAphyBphyC mutants, and measurement of the plant hormone levels indicated low ethylene production and bioactive GA levels in the phyAphyBphyC mutants
- PHYB~OsphyB, PHYC, The multiple contributions of phytochromes to the control of internode elongation in rice, We demonstrate that ethylene induced internode elongation and ACO1 expression in phyAphyBphyC seedlings but not in the wild type and that the presence of bioactive GAs was necessary for these effects
- PHYA~OsPhyA, PHYB~OsphyB, The multiple contributions of phytochromes to the control of internode elongation in rice, Although phyAphyBphyC phytochrome-null mutants in rice (Oryza sativa) have morphological changes and exhibit internode elongation, even as seedlings, it is unknown how phytochromes contribute to the control of internode elongation
- PHYA~OsPhyA, PHYB~OsphyB, The multiple contributions of phytochromes to the control of internode elongation in rice, A gene for 1-aminocyclopropane-1-carboxylate oxidase (ACO1), which is an ethylene biosynthesis gene contributing to internode elongation, was up-regulated in phyAphyBphyC seedlings
- PHYA~OsPhyA, PHYB~OsphyB, The multiple contributions of phytochromes to the control of internode elongation in rice, ACO1 expression was controlled mainly by phyA and phyB, and a histochemical analysis showed that ACO1 expression was localized to the basal parts of leaf sheaths of phyAphyBphyC seedlings, similar to mature wild-type plants at the heading stage, when internode elongation was greatly promoted
- PHYA~OsPhyA, PHYB~OsphyB, The multiple contributions of phytochromes to the control of internode elongation in rice, In addition, the transcription levels of several ethylene- or gibberellin (GA)-related genes were changed in phyAphyBphyC mutants, and measurement of the plant hormone levels indicated low ethylene production and bioactive GA levels in the phyAphyBphyC mutants
- PHYA~OsPhyA, PHYB~OsphyB, The multiple contributions of phytochromes to the control of internode elongation in rice, We demonstrate that ethylene induced internode elongation and ACO1 expression in phyAphyBphyC seedlings but not in the wild type and that the presence of bioactive GAs was necessary for these effects
- Ghd7, PHYB~OsphyB, A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice, We propose that both positive (OsMADS50 and Ehd1) and negative (Hd1, phyB and Ghd7) regulators of RFT1 form a gene network that regulates LD flowering in rice
- Hd1, PHYB~OsphyB, A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice, We propose that both positive (OsMADS50 and Ehd1) and negative (Hd1, phyB and Ghd7) regulators of RFT1 form a gene network that regulates LD flowering in rice
- OsMADS50~OsSOC1~DTH3, PHYB~OsphyB, A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice, We propose that both positive (OsMADS50 and Ehd1) and negative (Hd1, phyB and Ghd7) regulators of RFT1 form a gene network that regulates LD flowering in rice
- PHYB~OsphyB, RFT1, A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice, We propose that both positive (OsMADS50 and Ehd1) and negative (Hd1, phyB and Ghd7) regulators of RFT1 form a gene network that regulates LD flowering in rice
- Ehd1, PHYB~OsphyB, A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice, We propose that both positive (OsMADS50 and Ehd1) and negative (Hd1, phyB and Ghd7) regulators of RFT1 form a gene network that regulates LD flowering in rice
- OsJar1~OsGH3.5~OsGH3-5, PHYB~OsphyB, Rice JASMONATE RESISTANT 1 is involved in phytochrome and jasmonate signalling, The analysis of OsJar1 expression in phytochrome (phy) mutants revealed that phytochrome A (phyA) and phytochrome B (phyB) act redundantly to induce this gene by red light, presumably
- PHYA~OsPhyA, PHYB~OsphyB, Rice JASMONATE RESISTANT 1 is involved in phytochrome and jasmonate signalling, The analysis of OsJar1 expression in phytochrome (phy) mutants revealed that phytochrome A (phyA) and phytochrome B (phyB) act redundantly to induce this gene by red light, presumably
- Ehd1, PHYB~OsphyB, OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB, These pathways are: (i) night break-sensitive, which does not need OsCOL4; and (ii) night break-insensitive, in which OsCOL4 functions between OsphyB and Ehd1
- Ehd1, PHYB~OsphyB, OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB, OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB
- OsCOL4, PHYB~OsphyB, OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB, In osphyB mutants, OsCOL4 expression was decreased and osphyB oscol4 double mutants flowered at the same time as the osphyB single mutants, indicating OsCOL4 functions downstream of OsphyB
- OsCOL4, PHYB~OsphyB, OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB, These pathways are: (i) night break-sensitive, which does not need OsCOL4; and (ii) night break-insensitive, in which OsCOL4 functions between OsphyB and Ehd1
- OsCOL4, PHYB~OsphyB, OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB, OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB
- Hd1, PHYB~OsphyB, Footprints of natural and artificial selection for photoperiod pathway genes in Oryza, We examined the footprints of natural and artificial selections for four major genes of the photoperiod pathway, namely PHYTOCHROME B (PhyB), HEADING DATE 1 (Hd1), HEADING DATE 3a (Hd3a), and EARLY HEADING DATE 1 (Ehd1), by investigation of the patterns of nucleotide polymorphisms in cultivated and wild rice
- Ehd1, PHYB~OsphyB, Footprints of natural and artificial selection for photoperiod pathway genes in Oryza, We examined the footprints of natural and artificial selections for four major genes of the photoperiod pathway, namely PHYTOCHROME B (PhyB), HEADING DATE 1 (Hd1), HEADING DATE 3a (Hd3a), and EARLY HEADING DATE 1 (Ehd1), by investigation of the patterns of nucleotide polymorphisms in cultivated and wild rice
- Hd3a, PHYB~OsphyB, Footprints of natural and artificial selection for photoperiod pathway genes in Oryza, We examined the footprints of natural and artificial selections for four major genes of the photoperiod pathway, namely PHYTOCHROME B (PhyB), HEADING DATE 1 (Hd1), HEADING DATE 3a (Hd3a), and EARLY HEADING DATE 1 (Ehd1), by investigation of the patterns of nucleotide polymorphisms in cultivated and wild rice
- PHYA~OsPhyA, PHYB~OsphyB, Phytochromes are the sole photoreceptors for perceiving red/far-red light in rice, To directly address this hypothesis, a phytochrome triple mutant (phyAphyBphyC) was generated in rice (Oryza sativa L
- PHYA~OsPhyA, PHYB~OsphyB, Phytochromes are the sole photoreceptors for perceiving red/far-red light in rice, Since rice only has three phytochrome genes (PHYA, PHYB and PHYC), this mutant is completely lacking any phytochrome
- PHYB~OsphyB, PHYC, Phytochromes are the sole photoreceptors for perceiving red/far-red light in rice, To directly address this hypothesis, a phytochrome triple mutant (phyAphyBphyC) was generated in rice (Oryza sativa L
- PHYB~OsphyB, PHYC, Phytochromes are the sole photoreceptors for perceiving red/far-red light in rice, Since rice only has three phytochrome genes (PHYA, PHYB and PHYC), this mutant is completely lacking any phytochrome
- PHYA~OsPhyA, PHYB~OsphyB, Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice, The responses to Rc were completely canceled in phyA phyB double mutants
- PHYA~OsPhyA, PHYB~OsphyB, Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice, These results indicate that phyA and phyB act in a highly redundant manner to control deetiolation under Rc
- PHYA~OsPhyA, PHYB~OsphyB, Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice, Interestingly, the phyB phyC double mutant displayed clear R/FR reversibility in the pulse irradiation experiments, indicating that both phyA and phyB can mediate the low-fluence response for gene expression
- PHYA~OsPhyA, PHYB~OsphyB, Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice, Rice is a short-day plant, and we found that mutation in either phyB or phyC caused moderate early flowering under the long-day photoperiod, while monogenic phyA mutation had little effect on the flowering time
- PHYA~OsPhyA, PHYB~OsphyB, Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice, The phyA mutation, however, in combination with phyB or phyC mutation caused dramatic early flowering
- PHYB~OsphyB, PHYC, Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice, We have isolated phytochrome B (phyB) and phyC mutants from rice (Oryza sativa) and have produced all combinations of double mutants
- PHYB~OsphyB, PHYC, Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice, Seedlings of phyB and phyB phyC mutants exhibited a partial loss of sensitivity to continuous red light (Rc) but still showed significant deetiolation responses
- PHYB~OsphyB, PHYC, Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice, Interestingly, the phyB phyC double mutant displayed clear R/FR reversibility in the pulse irradiation experiments, indicating that both phyA and phyB can mediate the low-fluence response for gene expression
- PHYB~OsphyB, PHYC, Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice, Rice is a short-day plant, and we found that mutation in either phyB or phyC caused moderate early flowering under the long-day photoperiod, while monogenic phyA mutation had little effect on the flowering time
- PHYB~OsphyB, PHYC, Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice, The phyA mutation, however, in combination with phyB or phyC mutation caused dramatic early flowering
- PHYA~OsPhyA, PHYB~OsphyB, Comprehensive metabolite profiling of phyA phyB phyC triple mutants to reveal their associated metabolic phenotype in rice leaves, Rice (Oryza sativa) possesses three phytochromes, phyA, phyB, and phyC
- PHYA~OsPhyA, PHYB~OsphyB, Comprehensive metabolite profiling of phyA phyB phyC triple mutants to reveal their associated metabolic phenotype in rice leaves, In the present study, non-targeted metabolite analysis by gas chromatography time-of-flight mass spectrometry (GC/TOF-MS) and targeted metabolite analysis by capillary electrophoresis electrospray ionization mass spectrometry (CE/ESI-MS) were employed to investigate metabolic changes in rice phyA phyB phyC triple mutants
- PHYA~OsPhyA, PHYB~OsphyB, Comprehensive metabolite profiling of phyA phyB phyC triple mutants to reveal their associated metabolic phenotype in rice leaves, Distinct metabolic profiles between phyA phyB phyC triple mutants and the wild type (WT), as well as those between young and mature leaf blades, could be clearly observed by principal component analysis (PCA)
- PHYA~OsPhyA, PHYB~OsphyB, Comprehensive metabolite profiling of phyA phyB phyC triple mutants to reveal their associated metabolic phenotype in rice leaves, The metabolite profiles indicated high accumulation of amino acids, organic acids, sugars, sugar phosphates, and nucleotides in the leaf blades of phyA phyB phyC triple mutants, especially in the young leaves, compared with those in the WT
- PHYA~OsPhyA, PHYB~OsphyB, Comprehensive metabolite profiling of phyA phyB phyC triple mutants to reveal their associated metabolic phenotype in rice leaves, 5-fold), was observed in young leaves of phyA phyB phyC triple mutants
- PHYA~OsPhyA, PHYB~OsphyB, Comprehensive metabolite profiling of phyA phyB phyC triple mutants to reveal their associated metabolic phenotype in rice leaves, These metabolic phenotypes suggest that sugar metabolism, carbon partitioning, sugar transport, or some combination of these is impaired in the phyA phyB phyC triple mutants, and conversely, that phytochromes have crucial roles in sugar metabolism
- PHYA~OsPhyA, PHYB~OsphyB, Comprehensive metabolite profiling of phyA phyB phyC triple mutants to reveal their associated metabolic phenotype in rice leaves, Comprehensive metabolite profiling of phyA phyB phyC triple mutants to reveal their associated metabolic phenotype in rice leaves
- PHYB~OsphyB, PHYC, Comprehensive metabolite profiling of phyA phyB phyC triple mutants to reveal their associated metabolic phenotype in rice leaves, Rice (Oryza sativa) possesses three phytochromes, phyA, phyB, and phyC
- PHYB~OsphyB, PHYC, Comprehensive metabolite profiling of phyA phyB phyC triple mutants to reveal their associated metabolic phenotype in rice leaves, In the present study, non-targeted metabolite analysis by gas chromatography time-of-flight mass spectrometry (GC/TOF-MS) and targeted metabolite analysis by capillary electrophoresis electrospray ionization mass spectrometry (CE/ESI-MS) were employed to investigate metabolic changes in rice phyA phyB phyC triple mutants
- PHYB~OsphyB, PHYC, Comprehensive metabolite profiling of phyA phyB phyC triple mutants to reveal their associated metabolic phenotype in rice leaves, Distinct metabolic profiles between phyA phyB phyC triple mutants and the wild type (WT), as well as those between young and mature leaf blades, could be clearly observed by principal component analysis (PCA)
- PHYB~OsphyB, PHYC, Comprehensive metabolite profiling of phyA phyB phyC triple mutants to reveal their associated metabolic phenotype in rice leaves, The metabolite profiles indicated high accumulation of amino acids, organic acids, sugars, sugar phosphates, and nucleotides in the leaf blades of phyA phyB phyC triple mutants, especially in the young leaves, compared with those in the WT
- PHYB~OsphyB, PHYC, Comprehensive metabolite profiling of phyA phyB phyC triple mutants to reveal their associated metabolic phenotype in rice leaves, 5-fold), was observed in young leaves of phyA phyB phyC triple mutants
- PHYB~OsphyB, PHYC, Comprehensive metabolite profiling of phyA phyB phyC triple mutants to reveal their associated metabolic phenotype in rice leaves, These metabolic phenotypes suggest that sugar metabolism, carbon partitioning, sugar transport, or some combination of these is impaired in the phyA phyB phyC triple mutants, and conversely, that phytochromes have crucial roles in sugar metabolism
- PHYB~OsphyB, PHYC, Comprehensive metabolite profiling of phyA phyB phyC triple mutants to reveal their associated metabolic phenotype in rice leaves, Comprehensive metabolite profiling of phyA phyB phyC triple mutants to reveal their associated metabolic phenotype in rice leaves
- PHYA~OsPhyA, PHYB~OsphyB, OsPhyA modulates rice flowering time mainly through OsGI under short days and Ghd7 under long days in the absence of phytochrome B., We investigated the role of OsPhyA by comparing the osphyA osphyB double mutant to an osphyB single mutant
- PHYA~OsPhyA, PHYB~OsphyB, OsPhyA modulates rice flowering time mainly through OsGI under short days and Ghd7 under long days in the absence of phytochrome B., We also demonstrated that far-red light delays flowering time via both OsPhyA and OsPhyB
- OsCOL13, PHYB~OsphyB, A CONSTANS-like transcriptional activator, OsCOL13, functions as a negative regulator of flowering downstream of OsphyB and upstream of Ehd1 in rice., A CONSTANS-like transcriptional activator, OsCOL13, functions as a negative regulator of flowering downstream of OsphyB and upstream of Ehd1 in rice.
- OsCOL13, PHYB~OsphyB, A CONSTANS-like transcriptional activator, OsCOL13, functions as a negative regulator of flowering downstream of OsphyB and upstream of Ehd1 in rice., In addition, the transcriptional level of OsCOL13 significantly decreased in the osphyb mutant, but remained unchanged in the osphya and osphyc mutants
- OsCOL13, PHYB~OsphyB, A CONSTANS-like transcriptional activator, OsCOL13, functions as a negative regulator of flowering downstream of OsphyB and upstream of Ehd1 in rice., Thus, we conclude that OsCOL13 functions as a negative regulator downstream of OsphyB and upstream of Ehd1 in the photoperiodic flowering in rice
- APG~OsPIL16, PHYB~OsphyB, Phytochrome B Negatively Affects Cold Tolerance by Regulating OsDREB1 Gene Expression through Phytochrome Interacting Factor-Like Protein OsPIL16 in Rice., Expression pattern analyses revealed that OsPIL16 transcripts were induced by cold stress and was significantly higher in the phyB mutant than in the WT
- APG~OsPIL16, PHYB~OsphyB, Phytochrome B Negatively Affects Cold Tolerance by Regulating OsDREB1 Gene Expression through Phytochrome Interacting Factor-Like Protein OsPIL16 in Rice., Moreover, yeast two-hybrid assay showed that OsPIL16 can bind to rice PHYB
- APG~OsPIL16, PHYB~OsphyB, Phytochrome B Negatively Affects Cold Tolerance by Regulating OsDREB1 Gene Expression through Phytochrome Interacting Factor-Like Protein OsPIL16 in Rice., Based on these results, we propose that phyB deficiency positively regulates OsDREB1 expression through OsPIL16 to enhance cell membrane integrity and to reduce the malondialdehyde concentration, resulting in the improved cold tolerance of the phyB mutants