- Information
- Symbol: PNZIP,OsCRD1,YL-1
- MSU: LOC_Os01g17170
- RAPdb: Os01g0279100
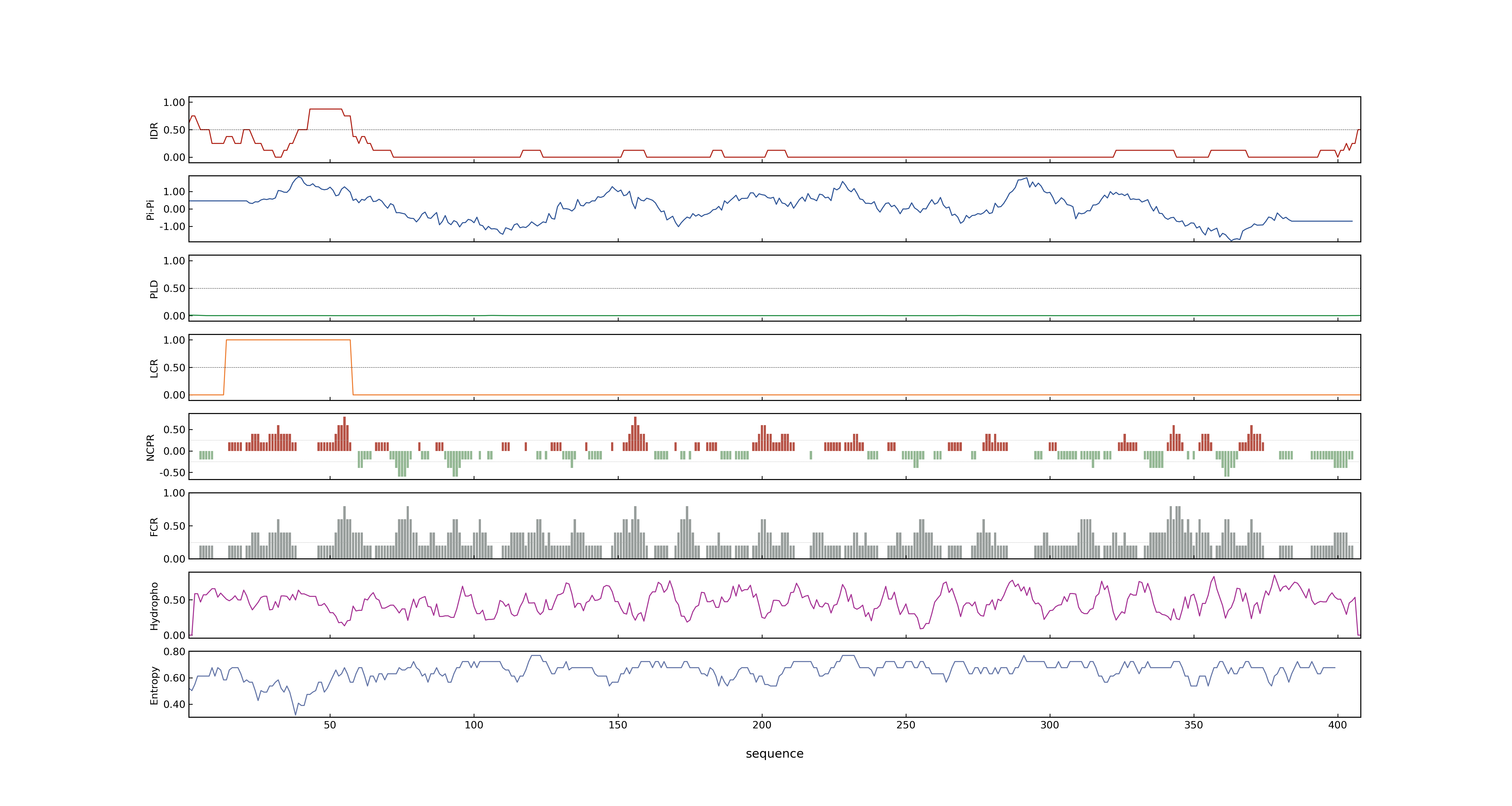
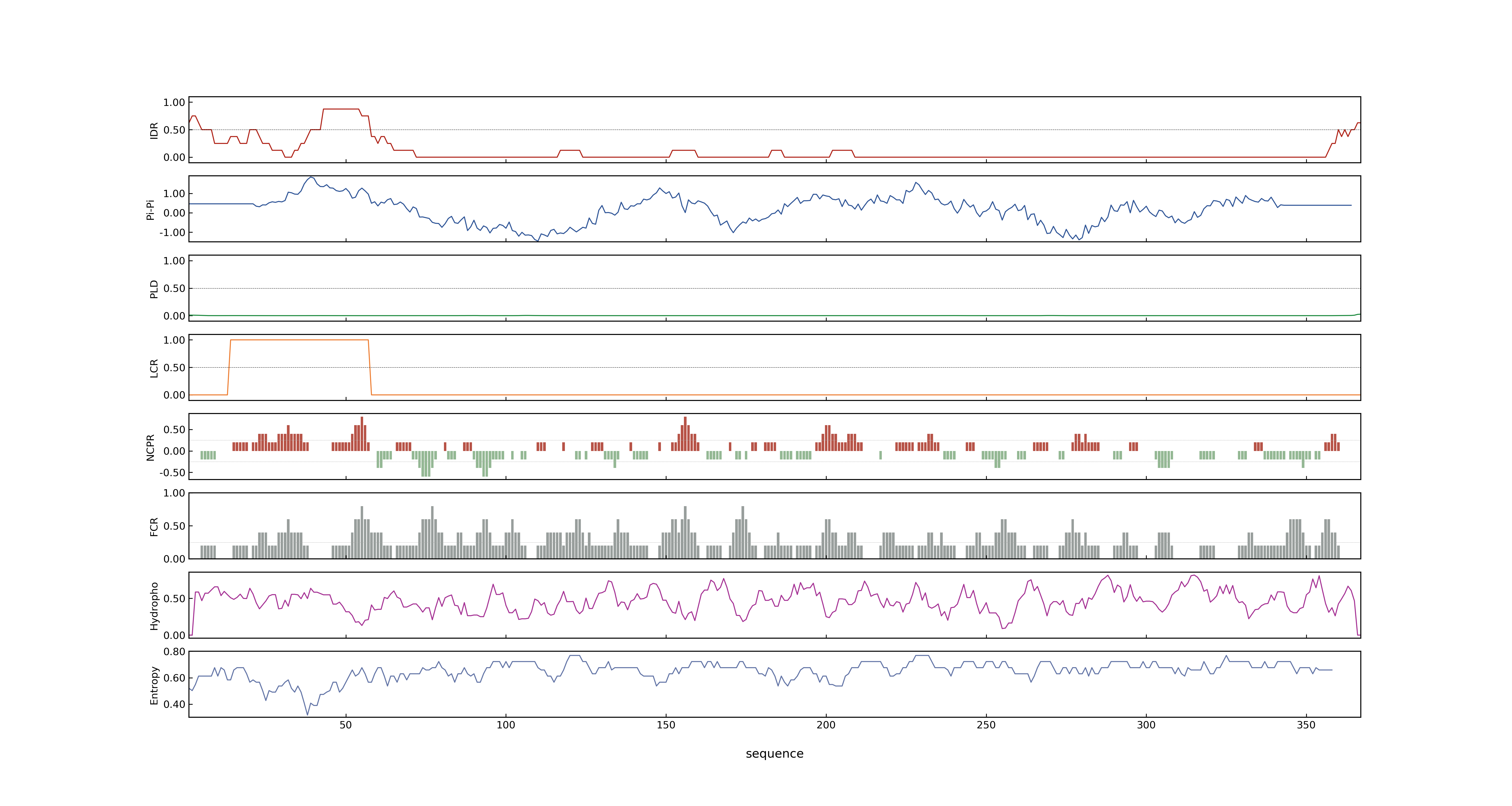
- PSP score
- LOC_Os01g17170.1: 0.0103
- LOC_Os01g17170.2: 0.0458
- PLAAC score
- LOC_Os01g17170.1: 0
- LOC_Os01g17170.2: 0
- pLDDT score
- NA
- MolPhase score
- LOC_Os01g17170.1: 0.01385631
- LOC_Os01g17170.2: 0.20481371
- MolPhase Result
- Publication
- Dissection of a QTL reveals an adaptive, interacting gene complex associated with transgressive variation for flowering time in rice, 2010, Theor Appl Genet.
- Substitution mapping of dth1.1, a flowering-time quantitative trait locus QTL associated with transgressive variation in rice, reveals multiple sub-QTL, 2006, Genetics.
- Yellow-Leaf 1 encodes a magnesium-protoporphyrin IX monomethyl ester cyclase, involved in chlorophyll biosynthesis in rice Oryza sativa L.., 2017, PLoS One.
- Genbank accession number
- Key message
- 1 region were associated with the presence of one or more flowering time genes (GI, SOC1, FT-L8, EMF1, and PNZIP)
- Under short-day lengths, lines with introgressions carrying combinations of linked flowering time genes (GI/SOC1, SOC1/FT-L8, GI/SOC1/FT-L8 and EMF1/PNZIP) from the late parent, O
- rufipogon, flowered earlier than the recurrent parent, Jefferson, while recombinant lines carrying smaller introgressions marked by the presence of GI, SOC1, EMF1 or PNZIP alone no longer flowered early
- Under long-day length, lines carrying SOC1/FT-L8, SOC1 or PNZIP flowered early, while those carrying GI or EMF1 delayed flowering
- Map-based cloning and over-expression analysis suggested that YL-1 encodes a subunit of MPEC
- Results of qRT-PCR showed that Chl biosynthesis upstream genes were highly expressed in the yl-1 mutant, while downstream genes were compromised, indicating that YL-1 plays a pivotal role in the Chl biosynthesis
- Connection
Prev Next