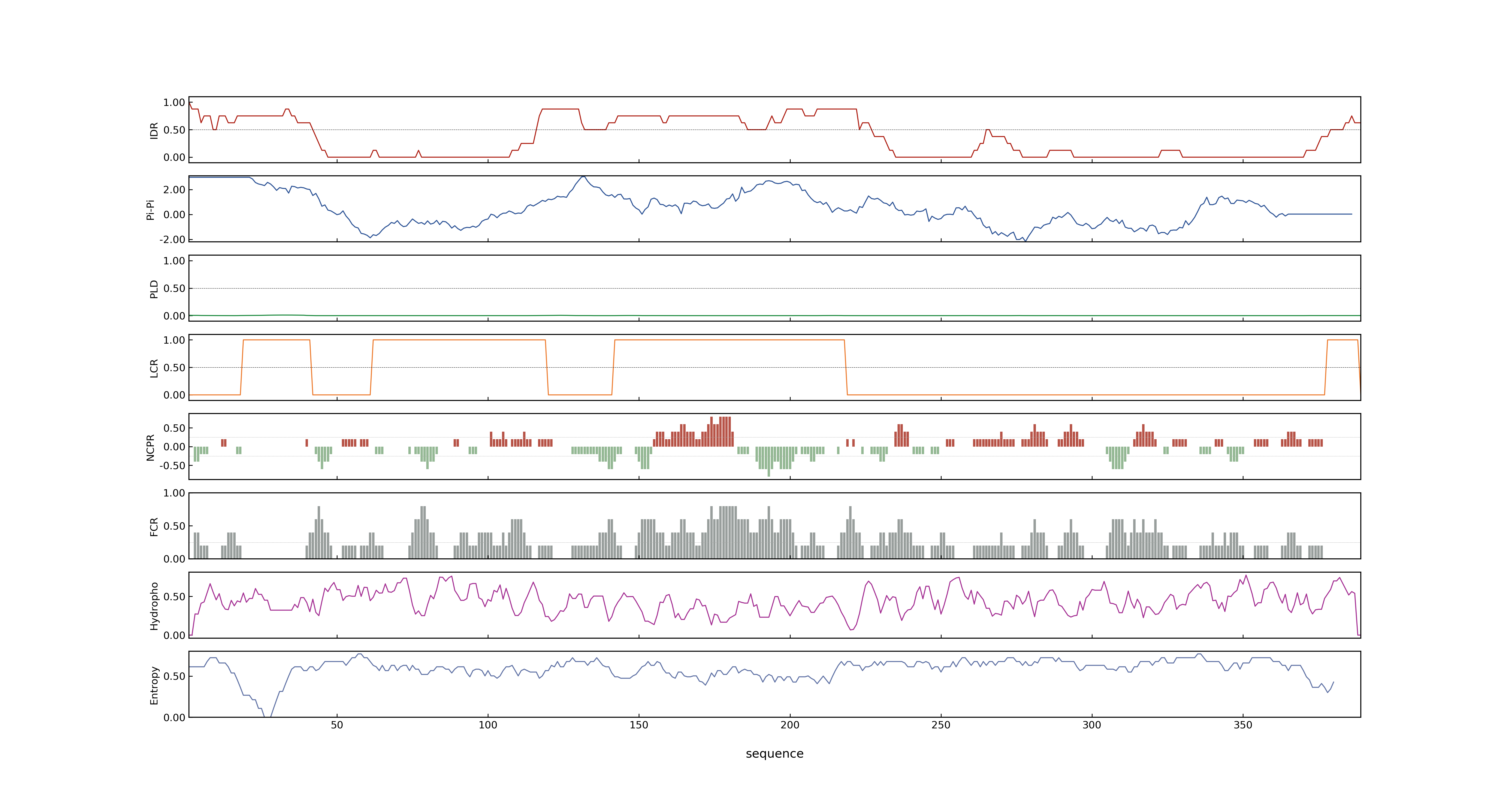
- Information
- Symbol: RFL,APO2,SSC
- MSU: LOC_Os04g51000
- RAPdb: Os04g0598300
- PSP score
- LOC_Os04g51000.1: 0.4762
- PLAAC score
- LOC_Os04g51000.1: 0
- pLDDT score
- NA
- MolPhase score
- LOC_Os04g51000.1: 0.99927276
- MolPhase Result
- Publication
- ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1, 2012, Plant J.
- Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture, 2008, Proc Natl Acad Sci U S A.
- Down-regulation of RFL, the FLO/LFY homolog of rice, accompanied with panicle branch initiation, 1998, Proc Natl Acad Sci U S A.
- Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture, 2008, Proceedings of the National Academy of Sciences.
- Dwarf and deformed flower 1, encoding an F-box protein, is critical for vegetative and floral development in rice Oryza sativa L., 2012, Plant J.
- Functions for rice RFL in vegetative axillary meristem specification and outgrowth., 2015, J Exp Bot.
- The far-upstream regulatory region of RFL is required for its precise spatial-temporal expression for floral development in rice., 2016, Plant Mol Biol.
- Short and Solid Culm/RFL/APO2 for culm development in rice., 2017, Plant J.
- Genbank accession number
- Key message
- Our data provide mechanistic insights into a unique role for RFL in determining the typical rice plant architecture by regulating distinct downstream pathways
- These results offer a means to alter rice flowering time and plant architecture by manipulating RFL-mediated pathways
- Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture
- To better understand the molecular mechanisms that regulate inflorescence and flower architecture, we characterized the rice aberrant panicle organization 2 (apo2) mutant which exhibits small panicles with reduced number of primary branches due to the precocious formation of spikelet meristems
- The apo2 mutants also display a shortened plastochron in the vegetative phase, late flowering, aberrant floral organ identities and loss of floral meristem determinacy
- Further analysis indicated that APO2/RFL and APO1, the rice ortholog of Arabidopsis UNUSUAL FLORAL ORGANS, act cooperatively to control inflorescence and flower development
- Aside from facilitating a transition of the main growth axis to an inflorescence meristem, RFL expression status affects vegetative axillary meristems and therefore regulates tillering
- The unique spatially and temporally regulated RFL expression during the development of vegetative axillary bud (tiller) primordia and inflorescence branch primordia is therefore required to produce tillers and panicle branches, respectively
- Diverse orthologous LFY genes regulate vegetative-to-reproductive phase transition when expressed in Arabidopsis, a property not shared by RFL, the homolog in the agronomically important grass, rice
- After the transition to reproductive stage, RFL RNA was detected in all layers of very young panicle including the apical meristem, but absent in the incipient primary branches
- ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1
- In particular, APO2/RFL and LFY act oppositely on inflorescence development
- Northern analysis showed that RFL is expressed predominantly in very young panicle but not in mature florets, mature leaves, or roots
- Expression pattern of RFL raised a possibility that, unlike FLO and LFY, RFL might be involved in panicle branching
- Down-regulation of RFL, the FLO/LFY homolog of rice, accompanied with panicle branch initiation
- In situ hybridization revealed that RFL RNA was expressed in epidermal cells in young leaves at vegetative growth stage
- Map-based cloning revealed that APO2 is identical to previously reported RFL gene, the rice ortholog of the Arabidopsis LEAFY (LFY) gene
- In these transgenics, the expression levels of known flowering time genes reveal RFL as a regulator of OsSOC1 (OsMADS50), an activator of flowering
- We find that reduction in RFL expression causes a dramatic delay in transition to flowering, with the extreme phenotype being no flowering
- Conversely, RFL overexpression triggers precocious flowering
- In 35S-RFL plants, transformation of inflorescence meristem to floral meristem was rarely observed
- As development of branches proceeds, RFL RNA accumulation localized in the developing branches except for the apical meristems of the branches and secondary branch primordia
- Instead, development of cotyledons, rosette leaves, petals, and stamens was severely affected, demonstrating that RFL function is distinct from that of LFY
- To examine whether same mechanisms control floral development in distantly related species such as grasses, we isolated RFL, FLO-LFY homolog of rice, and examined its expression and function
- Transgenic Arabidopsis plants constitutively expressing RFL from the cauliflower mosaic virus 35S promoter were produced to test whether 35S-RFL would cause similar phenotype as observed in 35S-LFY plants
- Previous studies showed that knockdown of the transcription factor gene RFL reduced tillering and caused a very large decrease in panicle branching
- Functions for rice RFL in vegetative axillary meristem specification and outgrowth.
- Further, we report reduced expression of auxin transporter genes OsPIN1 and OsPIN3 in the culm of RFL knockdown transgenic plants
- Additionally, subtle change in the spatial pattern of IR4 DR5:GFP auxin reporter was observed, which hints at compromised auxin transport on RFL knockdown
- Together, we have found that RFL regulates the LAX1 and CUC genes during AM specification, and positively influences the outgrowth of AMs though its effects on auxin transport
- The relationship between RFL, strigolactone signalling, and bud outgrowth was studied by transcript analyses and by the tillering phenotype of transgenic plants knocked down for both RFL and D3
- The in situ hybridization result showed that, when florets appear, RFL was expressed almost exclusively at the palea/lemma adaxial base adjacent to lodicules in the wild-type panicle
- The far-upstream regulatory region of RFL is required for its precise spatial-temporal expression for floral development in rice.
- The results suggest that the far-upstream region of RFL may contain potential cis element(s) that are critical to define the precise spatial-temporal expression pattern of RFL for its function in floral development
- A disruption of sequence integrity at 6292-bp upstream of RFL by a T-DNA insertion led to varied RFL expression patterns in floral meristem and floret in afo1 and caused the mutant phenotype
- Here, we describe a rice aberrant floral organ 1 (afo1) mutant that was produced by a T-DNA insertion at 6292-bp upstream of the start codon of RFL
- This insertion altered the expression of RFL in floral meristem (FM) and floret
- The altered pattern of RFL expression in afo1 resulted in enlarged FMs, more floral organs, aberrant floral organ identity, and loss of FM determinacy
- Connection
- APO1~OsAPO1~SCM2, RFL~APO2~SSC, ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1, Further analysis indicated that APO2/RFL and APO1, the rice ortholog of Arabidopsis UNUSUAL FLORAL ORGANS, act cooperatively to control inflorescence and flower development
- APO1~OsAPO1~SCM2, RFL~APO2~SSC, ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1, ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1
- OsMADS50~OsSOC1~DTH3, RFL~APO2~SSC, Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture, In these transgenics, the expression levels of known flowering time genes reveal RFL as a regulator of OsSOC1 (OsMADS50), an activator of flowering
- DDF1, RFL~APO2~SSC, Dwarf and deformed flower 1, encoding an F-box protein, is critical for vegetative and floral development in rice Oryza sativa L., In addition, DDF1 also negatively regulates the Arabidopsis LFY ortholog APO2, implying a functional connection between DDF1 and APO2
- D3, RFL~APO2~SSC, Functions for rice RFL in vegetative axillary meristem specification and outgrowth., The relationship between RFL, strigolactone signalling, and bud outgrowth was studied by transcript analyses and by the tillering phenotype of transgenic plants knocked down for both RFL and D3
- LAX1, RFL~APO2~SSC, Functions for rice RFL in vegetative axillary meristem specification and outgrowth., We show that RFL promotes AM specification through its effects on LAX1 and CUC genes, as their expression was modulated on RFL knockdown, on induction of RFL:GR fusion protein, and by a repressive RFL-EAR fusion protein
- LAX1, RFL~APO2~SSC, Functions for rice RFL in vegetative axillary meristem specification and outgrowth., Together, we have found that RFL regulates the LAX1 and CUC genes during AM specification, and positively influences the outgrowth of AMs though its effects on auxin transport
- OsPIN1~REH1~PIN1b, RFL~APO2~SSC, Functions for rice RFL in vegetative axillary meristem specification and outgrowth., Further, we report reduced expression of auxin transporter genes OsPIN1 and OsPIN3 in the culm of RFL knockdown transgenic plants
- OsPIN1~REH1~PIN1b, RFL~APO2~SSC, Functions for rice RFL in vegetative axillary meristem specification and outgrowth., These modified forms of RFL had only indirect effects on OsPIN1
- FZP~BFL1, RFL~APO2~SSC, Regulatory role of FZP in the determination of panicle branching and spikelet formation in rice., FRIZZLE PANICLE (FZP) and RFL/ABERRANT PANICLE ORGANIZATION 2 (APO2) play important roles in regulating the ABCDE floral organ identity genes
Prev Next