- Information
- Symbol: Rcn1,OsABCG5
- MSU: LOC_Os03g17350
- RAPdb: Os03g0281900
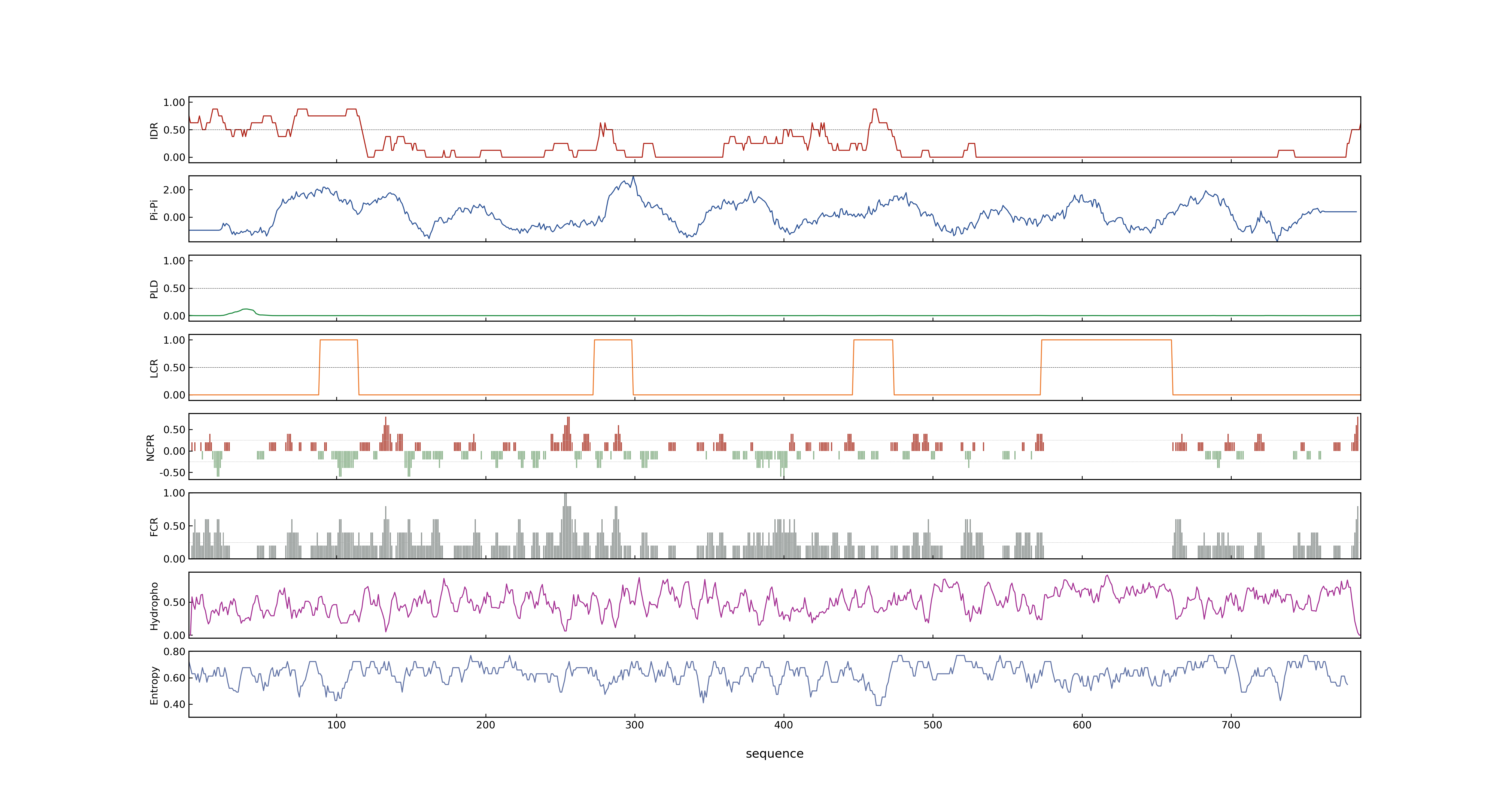
- PSP score
- LOC_Os03g17350.1: 0.5305
- PLAAC score
- LOC_Os03g17350.1: 0
- pLDDT score
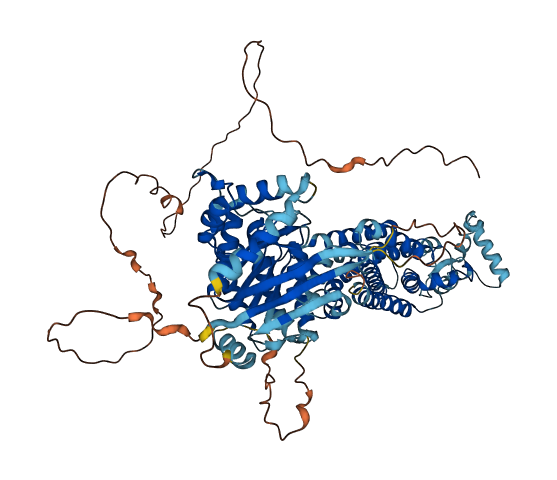
- 77.03
- Protein Structure from AlphaFold and UniProt
- MolPhase score
- LOC_Os03g17350.1: 0.99858653
- MolPhase Result
- Publication
- The rice REDUCED CULM NUMBER11 gene controls vegetative growth under low-temperature conditions in paddy fields independent of RCN1/OsABCG5, 2013, Plant Sci.
- Genetic interaction between 2 tillering genes, reduced culm number 1 rcn1 and tillering dwarf gene d3, in rice, 2007, J Hered.
- Overexpression ofRCN1andRCN2, riceTERMINAL FLOWER 1/CENTRORADIALIShomologs, confers delay of phase transition and altered panicle morphology in rice, 2002, The Plant Journal.
- Rice shoot branching requires an ATP-binding cassette subfamily G protein, 2009, New Phytol.
- The ethylene receptor ETR2 delays floral transition and affects starch accumulation in rice, 2009, Plant Cell.
- RCN1/OsABCG5, an ATP-binding cassette ABC transporter, is required for hypodermal suberization of roots in rice Oryza sativa., 2014, Plant J.
- Ethylene Biosynthesis is Promoted by Very-Long-Chain Fatty Acids during Lysigenous Aerenchyma Formation in Rice Roots., 2015, Plant Physiol.
- Rice stomatal closure requires guard cell plasma membrane ATP-binding cassette transporter RCN1/OsABCG5., 2015, Mol Plant.
-
Genbank accession number
- Key message
- To investigate the possibility of similar mechanisms operating in the control of inflorescence architecture in rice, we analysed the functions of RCN1 and RCN2, rice TFL1/CEN homologs
- Phenotypic analyses of rcn1 and tillering dwarf 3 (d3) double mutants at the seedling stage clarified that Rcn1 works independently of D3 in the branching inhibitor pathway
- Since the shoot architecture of the rcn11 was very similar to that of the rcn1, we examined whether RCN11 is involved in RCN1/OsABCG5-associated vegetative growth control
- The rcn1 rcn11 phenotype suggests that RCN11 acts on vegetative growth independent of RCN1/OsABCG5
- Thus, rcn11 will shed new light on vegetative growth control under low temperature
- The rice REDUCED CULM NUMBER11 gene controls vegetative growth under low-temperature conditions in paddy fields independent of RCN1/OsABCG5
- A root development comparison between rcn1 and rcn11 in young seedlings represented that rcn11 reduced crown root number and elongation, whereas rcn1 reduced lateral root density and elongation
- We isolated a novel reduced culm number mutant, designated reduced culm number11 (rcn11), by screening under low-temperature condition in a paddy fields
- In 35S::RCN1 and 35S::RCN2 transgenic rice plants, the delay of transition to the reproductive phase was observed
- Mutant genes, reduced culm number 1 (rcn1) and bunketsuwaito tillering dwarf (d3), affect tiller number in rice (Oryza sativa L
- Genetic interaction between 2 tillering genes, reduced culm number 1 (rcn1) and tillering dwarf gene d3, in rice
- In addition, Rcn1 is expressed in the crown root primordia, endodermis, pericycle and stele in the root
- A new rcn1 mutant, designated as S-97-61 exhibited a reduction in tiller number and plant stature to about the same level as the previously reported original rcn1 mutant
- The reduction in tillering by the rcn1 mutation was independent of the d3 genotype, and tillering number of d3rcn1 double mutant was between those of the d3 and rcn1 mutants
- These results demonstrated that the Rcn1 gene was not involved in the D3-associated pathway in tillering control
- The GIGANTEA and TERMINAL FLOWER1/CENTRORADIALIS homolog (RCN1) that cause delayed flowering were upregulated in ETR2-overexpressing plants but downregulated in the etr2 mutant
- Constitutive overexpression of RCN1 or RCN2 in Arabidopsis caused a late-flowering and highly branching phenotype, indicating that they possess conserved biochemical functions as TFL1
- No effect on Rcn1 expression in shoots or roots was seen when the roots were treated with auxins
-
- Here, characterization of rice (Oryza sativa) reduced culm number 1 (rcn1) mutants revealed that Rcn1 positively controls shoot branching by promoting the outgrowth of lateral shoots
- Overexpression ofRCN1andRCN2, riceTERMINAL FLOWER 1/CENTRORADIALIShomologs, confers delay of phase transition and altered panicle morphology in rice
-
- Rcn1 is the first functionally defined plant ABCG protein gene that controls shoot branching and could thus be significant in future breeding for high-yielding rice
-
- Rcn1 is expressed in leaf primordia of main and axillary shoots, and in the vascular cells and leaf epidermis of older leaves
- RCN1/OsABCG5, an ATP-binding cassette (ABC) transporter, is required for hypodermal suberization of roots in rice (Oryza sativa).
- In many plant species, including rice (Oryza sativa), the hypodermis in the outer part of roots forms a suberized cell wall (the Casparian strip and/or suberin lamellae), which inhibits the flow of water and ions and protects against pathogens
- We discovered that a rice reduced culm number1 (rcn1) mutant could not develop roots longer than 100 mm in waterlogged soil
- RCN1/OsABCG5 gene expression in the wild type was increased in most hypodermal and some endodermal roots cells under stagnant deoxygenated conditions
- These findings suggest that RCN1/OsABCG5 has a role in the suberization of the hypodermis of rice roots, which contributes to formation of the apoplastic barrier
- The mutated gene encoded an ATP-binding cassette (ABC) transporter named RCN1/OsABCG5
- A GFP-RCN1/OsABCG5 fusion protein localized at the plasma membrane of the wild type
- ABA application resulted in a smaller increase in the percentage of guard cell pairs containing ABA in rcn1 mutant (A684P) and RCN1-RNAi than in wild-type plants
- Furthermore, polyethylene glycol (drought stress)-inducible ABA accumulation in guard cells did not occur in rcn1 mutants
- Stomata closure mediated by exogenous ABA application was strongly reduced in rcn1 mutants
- Finally, rcn1 mutant plants had more rapid water loss from detached leaves than the wild-type plants
- Connection
- D3, Rcn1~OsABCG5, Genetic interaction between 2 tillering genes, reduced culm number 1 rcn1 and tillering dwarf gene d3, in rice, Mutant genes, reduced culm number 1 (rcn1) and bunketsuwaito tillering dwarf (d3), affect tiller number in rice (Oryza sativa L
- D3, Rcn1~OsABCG5, Genetic interaction between 2 tillering genes, reduced culm number 1 rcn1 and tillering dwarf gene d3, in rice, Our objective was to compare the phenotype of the d3rcn1 double mutant with each single mutant and parental rice cultivar “Shiokari” and to clarify whether the Rcn1 gene interacted with the D3 gene
- D3, Rcn1~OsABCG5, Genetic interaction between 2 tillering genes, reduced culm number 1 rcn1 and tillering dwarf gene d3, in rice, We recovered a new rcn1 mutant from Shiokari and developed d3rcn1 double mutant with Shiokari genetic background
- D3, Rcn1~OsABCG5, Genetic interaction between 2 tillering genes, reduced culm number 1 rcn1 and tillering dwarf gene d3, in rice, Three near-isogenic lines, rcn1 mutant, d3 mutant, and d3rcn1 double mutant, were grown together with the parental Shiokari
- D3, Rcn1~OsABCG5, Genetic interaction between 2 tillering genes, reduced culm number 1 rcn1 and tillering dwarf gene d3, in rice, The reduction in tillering by the rcn1 mutation was independent of the d3 genotype, and tillering number of d3rcn1 double mutant was between those of the d3 and rcn1 mutants
- D3, Rcn1~OsABCG5, Genetic interaction between 2 tillering genes, reduced culm number 1 rcn1 and tillering dwarf gene d3, in rice, These results demonstrated that the Rcn1 gene was not involved in the D3-associated pathway in tillering control
- D3, Rcn1~OsABCG5, Genetic interaction between 2 tillering genes, reduced culm number 1 rcn1 and tillering dwarf gene d3, in rice, Genetic interaction between 2 tillering genes, reduced culm number 1 (rcn1) and tillering dwarf gene d3, in rice
- Rcn1~OsABCG5, RCN2, Overexpression ofRCN1andRCN2, riceTERMINAL FLOWER 1/CENTRORADIALIShomologs, confers delay of phase transition and altered panicle morphology in rice, To investigate the possibility of similar mechanisms operating in the control of inflorescence architecture in rice, we analysed the functions of RCN1 and RCN2, rice TFL1/CEN homologs
- Rcn1~OsABCG5, RCN2, Overexpression ofRCN1andRCN2, riceTERMINAL FLOWER 1/CENTRORADIALIShomologs, confers delay of phase transition and altered panicle morphology in rice, Constitutive overexpression of RCN1 or RCN2 in Arabidopsis caused a late-flowering and highly branching phenotype, indicating that they possess conserved biochemical functions as TFL1
- Rcn1~OsABCG5, RCN2, Overexpression ofRCN1andRCN2, riceTERMINAL FLOWER 1/CENTRORADIALIShomologs, confers delay of phase transition and altered panicle morphology in rice, In 35S::RCN1 and 35S::RCN2 transgenic rice plants, the delay of transition to the reproductive phase was observed
- Rcn1~OsABCG5, RCN2, Overexpression ofRCN1andRCN2, riceTERMINAL FLOWER 1/CENTRORADIALIShomologs, confers delay of phase transition and altered panicle morphology in rice, Overexpression ofRCN1andRCN2, riceTERMINAL FLOWER 1/CENTRORADIALIShomologs, confers delay of phase transition and altered panicle morphology in rice
- ETR2~Os-ERL1~OsETR2, Rcn1~OsABCG5, The ethylene receptor ETR2 delays floral transition and affects starch accumulation in rice, The GIGANTEA and TERMINAL FLOWER1/CENTRORADIALIS homolog (RCN1) that cause delayed flowering were upregulated in ETR2-overexpressing plants but downregulated in the etr2 mutant
- OsXylT~RCN11, Rcn1~OsABCG5, The rice REDUCED CULM NUMBER11 gene controls vegetative growth under low-temperature conditions in paddy fields independent of RCN1/OsABCG5., Since the shoot architecture of the rcn11 was very similar to that of the rcn1, we examined whether RCN11 is involved in RCN1/OsABCG5-associated vegetative growth control
- OsXylT~RCN11, Rcn1~OsABCG5, The rice REDUCED CULM NUMBER11 gene controls vegetative growth under low-temperature conditions in paddy fields independent of RCN1/OsABCG5., A genetic allelism test and molecular mapping study showed that rcn11 is independent of rcn1 on rice chromosome 3 and located on chromosome 8
- OsXylT~RCN11, Rcn1~OsABCG5, The rice REDUCED CULM NUMBER11 gene controls vegetative growth under low-temperature conditions in paddy fields independent of RCN1/OsABCG5., The rcn1 rcn11 phenotype suggests that RCN11 acts on vegetative growth independent of RCN1/OsABCG5
- OsXylT~RCN11, Rcn1~OsABCG5, The rice REDUCED CULM NUMBER11 gene controls vegetative growth under low-temperature conditions in paddy fields independent of RCN1/OsABCG5., A root development comparison between rcn1 and rcn11 in young seedlings represented that rcn11 reduced crown root number and elongation, whereas rcn1 reduced lateral root density and elongation
Prev Next